Processing with OCSSW Command Line Interface (CLI)#
Tutorial Lead: Carina Poulin (NASA, SSAI)
The following notebooks are prerequisites for this tutorial:
Summary#
SeaDAS is the official data analysis sofware of NASA’s Ocean Biology Distributed Active Archive Center (OB.DAAC); used to process, display and analyse ocean color data. SeaDAS is a dektop application that includes the Ocean Color Science Software (OCSSW) libraries. There is also a command line interface (CLI) for the OCSSW libraries, which we can use to write processing scripts and notebooks.
This tutorial will show you how to process PACE OCI data using the sequence of OCSSW programs l2gen, l2bin, and l3mapgen.
Learning Objectives#
At the end of this notebok you will know:
How to process Level-1B data to Level-2 with
l2gento make different products including surface reflectances and MOANA phytoplankton productsHow to merge two images with
l2binHow to create a map with
l3mapgen
Contents#
1. Setup#
Begin by importing all of the packages used in this notebook. If your kernel uses an environment defined following the guidance on the tutorials page, then the imports will be successful.
import csv
import os
import cartopy.crs as ccrs
import earthaccess
import xarray as xr
import matplotlib.pyplot as plt
We are also going to define a function to help write OCSSW parameter files, which
is needed several times in this tutorial. To write the results in the format understood
by OCSSW, this function uses the csv.writer from the Python Standard Library. Instead of
writing comma-separated values, however, we specify a non-default delimiter to get
equals-separated values. Not something you usually see in a data file, but it’s better than
writing our own utility from scratch!
def write_par(path, par):
"""
Prepare a "par file" to be read by one of the OCSSW tools, as an
alternative to specifying each parameter on the command line.
Args:
path (str): where to write the parameter file
par (dict): the parameter names and values included in the file
"""
with open(path, "w") as file:
writer = csv.writer(file, delimiter="=")
values = writer.writerows(par.items())
The Python docstring (fenced by triple quotation marks in the function definition) is not essential, but it helps describe what the function does.
help(write_par)
Help on function write_par in module __main__:
write_par(path, par)
Prepare a "par file" to be read by one of the OCSSW tools, as an
alternative to specifying each parameter on the command line.
Args:
path (str): where to write the parameter file
par (dict): the parameter names and values included in the file
2. Get L1B Data#
Set (and persist to your user profile on the host, if needed) your Earthdata Login credentials.
auth = earthaccess.login(persist=True)
We will use the earthaccess search method used in the OCI Data Access notebook. Note that Level-1B (L1B) files
do not include cloud coverage metadata, so we cannot use that filter. In this search, the spatial filter is
performed on a location given as a point represented by a tuple of latitude and longitude in decimal degrees.
tspan = ("2024-06-05", "2024-06-05")
location = (40, 45)
results = earthaccess.search_data(
short_name="PACE_OCI_L1B_SCI",
temporal=tspan,
point=location,
)
results[0]
Download the granules found in the search.
paths = earthaccess.download(results, local_path="data")
While we have the downloaded location stored in the list paths, store one in a variable we won’t overwrite for future use.
l2gen_ifile = paths[0]
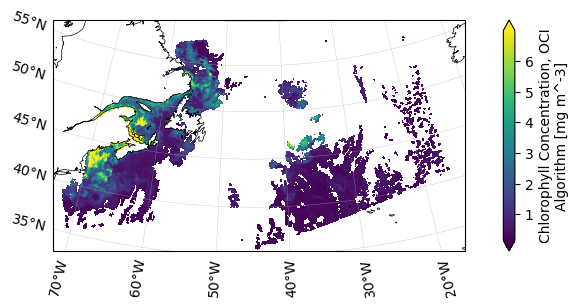
The Level-1B files contain top-of-atmosphere reflectances, typically denoted as \(\rho_t\).
On OCI, the reflectances are grouped into blue, red, and short-wave infrared (SWIR) wavelengths. Open
the dataset’s “observation_data” group in the netCDF file using xarray to plot a “rhot_red”
wavelength.
dataset = xr.open_dataset(l2gen_ifile, group="observation_data")
artist = dataset["rhot_red"].sel({"red_bands": 100}).plot()
3. Process L1B Data with l2gen#
At Level-1, we neither have geophysical variables nor are the data projected for easy map making. We will need to process the L1B file to Level-2 and then to Level-3 to get both of those. Note that Level-2 data for many geophysical variables are available for download from the OB.DAAC, so you often don’t need the first step. However, the Level-3 data distributed by the OB.DAAC are global composites, which may cover more Level-2 scenes than you want. You’ll learn more about compositing below. l2gen also allows you to customize products in a way that allows you to get products you cannot directly download from earthdata, like you will see from the following two examples. The first shows how to produce surface reflectances to make true-color images. The second covers the upcoming MOANA phytoplankton community composition products.
This section shows how to use l2gen for processing the L1B data to L2 using customizable parameters.
OCSSW programs are run from the command line in Bash, but we can have a Bash terminal-in-a-cell using the IPython magic command %%bash. In the specific case of OCSSW programs, the Bash environment created for that cell must be set up by loading $OCSSWROOT/OCSSW_bash.env.
Every %%bash cell that calls an OCSSW program needs to source the environment
definition file shipped with OCSSW, because its effects are not retained from one cell to the next.
We can, however, define the OCSSWROOT environment variable in a way that effects every %%bash cell.
os.environ.setdefault("OCSSWROOT", "/opt/ocssw")
'/opt/ocssw'
Then we need a couple lines, which will appear in multiple cells below, to begin a Bash cell initiated with the OCSSW_bash.env file.
Using this pattern, run the l2gen command with the single argument help to view the extensive list of options available. You can find more information about l2gen and other OCSSW functions on the seadas website
%%bash
source $OCSSWROOT/OCSSW_bash.env
l2gen help
msl12 9.7.0-V2024.1 (Jul 17 2024 13:15:09)
Usage: msl12 argument-list
The argument-list is a set of keyword=value pairs. The arguments can
be specified on the commandline, or put into a parameter file, or the
two methods can be used together, with commandline over-riding.
return value: 0=OK, 1=error, 110=north,south,east,west does not intersect
file data.
The list of valid keywords follows:
help (boolean) (alias=h) (default=false) = print usage information
version (boolean) (default=false) = print the version
information
dump_options (boolean) (default=false) = print
information about each option
dump_options_paramfile (ofile) = print
information about each option to paramfile
dump_options_xmlfile (ofile) = print
information about each option to XML file
par (ifile) = input parameter file
pversion (string) (default=Unspecified) = processing version string
suite (string) (default=OC) = product suite string for loading
suite-specific defaults
eval (int) (default=0) = evaluation bitmask
0: standard processing
1: init to old aerosol models
2: enables MODIS and MERIS cloud Mask for HABS
16: enables MODIS cirrus mask
32: use test sensor info file
64: use test rayleigh tables
128: use test aerosol tables
256: use test polarization tables
1024: mask modis mirror-side 1 (navfail)
2048: mask modis mirror-side 2 (navfail)
4096: don't apply 'cold-only' or equatorial aerosol tests for SST
8192: use alt sensor info file in eval
32768: enables spherical path geom for dtran
ifile (ifile) (alias=ifile1) = input L1 file name
ilist (ifile) = file containing list of input files, one per line
geofile (ifile) = input L1 geolocation file name (MODIS/VIIRS only)
ofile (ofile) (alias=ofile1) (default=output) = output file #1 name,
output vicarious L1B for inverse mode
ofile[#] = additional output L2 file name
oformat (string) (default=netCDF4) = output file format
netcdf4: output a netCDF version 4 file
hdf4: output a HDF version 4 file
il2file (ifile) (alias=il2file1) = input L2 file names for sensor to be
used as a calibrator. Alternatively, a data point can be used as a
calibrator (e.g. MOBY)
il2file[#] = additional L2 calibration file names
tgtfile (ifile) = vicarious calibration target file
aerfile (ifile) = aerosol model specification file
metafile (ifile) = output meta-data file
l2prod (string) (alias=l2prod1) = L2 products to be included in ofile #1
l2prod[#] = L2 products to be included in ofile[#]
proc_ocean (int) (default=1) = toggle ocean processing
1: On
0: Off
2: force all pixels to be processed as ocean
proc_land (boolean) (default=off) = toggle land processing
proc_sst (boolean) (default=false) = toggle SST processing
(default=1 for MODIS, 0 otherwise)
proc_cloud (boolean) (default=off) = toggle cloud processing
proc_uncertainty (int) (default=0) = uncertainty propagation mode
0: without uncertainty propagation
1: uncertainty propagation generating error variance
atmocor (boolean) (default=on) = toggle atmospheric correction
mode (int) (default=0) = processing mode
0: forward processing
1: inverse (calibration) mode, targeting to nLw=0
2: inverse (calibration) mode, given nLw target
3: inverse (calibration) mode, given Lw target (internally normalized)
aer_opt (int) (default=99) = aerosol mode option
-99: No aerosol subtraction
>0: Multi-scattering with fixed model (provide model number, 1-N,
relative to aermodels list)
0: White aerosol extrapolation.
-1: Multi-scattering with 2-band model selection
-2: Multi-scattering with 2-band, RH-based model selection and
iterative NIR correction
-3: Multi-scattering with 2-band model selection
and iterative NIR correction
-4: Multi-scattering with fixed model pair
(requires aermodmin, aermodmax, aermodrat specification)
-5: Multi-scattering with fixed model pair
and iterative NIR correction
(requires aermodmin, aermodmax, aermodrat specification)
-6: Multi-scattering with fixed angstrom
(requires aer_angstrom specification)
-7: Multi-scattering with fixed angstrom
and iterative NIR correction
(requires aer_angstrom specification)
-8: Multi-scattering with fixed aerosol optical thickness
(requires taua specification)
-9: Multi-scattering with 2-band model selection using Wang et al. 2009
to switch between SWIR and NIR. (MODIS only, requires aer_swir_short,
aer_swir_long, aer_wave_short, aer_wave_long)
-10: Multi-scattering with MUMM correction
and MUMM NIR calculation
-17: Multi-scattering epsilon, RH-based model selection
and iterative NIR correction
-18: Spectral Matching of aerosols reflectance
and iterative NIR correction
-19: Multi-scattering epsilon (linear), RH-based model selection
and iterative NIR correction
aer_wave_short (int) (default=765) = shortest sensor wavelength for aerosol
model selection
aer_wave_long (int) (default=865) = longest sensor wavelength for aerosol
model selection
aer_wave_base (int) (default=865) = base sensor wavelength for aerosol
extrapolation
aer_swir_short (int) (default=-1) = shortest sensor wavelength for
SWIR-based NIR Lw correction
aer_swir_long (int) (default=-1) = longest sensor wavelength for SWIR-based
NIR Lw correction
aer_rrs_short (float) (default=-1.0) = Rrs at shortest sensor wavelength for
aerosol model selection
aer_rrs_long (float) (default=-1.0) = Rrs at longest sensor wavelength for
aerosol model selection
aermodmin (int) (default=-1) = lower-bounding model to use for fixed model
pair aerosol option
aermodmax (int) (default=-1) = upper-bounding model to use for fixed model
pair aerosol option
aermodrat (float) (default=0.0) = ratio to use for fixed model pair aerosol
option
aer_angstrom (float) (default=-999.0) = aerosol angstrom exponent for model
selection
aer_iter_max (int) (default=10) = maximum number of iterations for NIR
water-leaving radiance estimation.
mumm_alpha (float) (default=1.72) = water-leaving reflectance ratio for MUMM
turbid water atmospheric correction
mumm_gamma (float) (default=1.0) = two-way Rayleigh-aerosol transmittance
ratio for MUMM turbid water atmospheric correction
mumm_epsilon (float) (default=1.0) = aerosol reflectance ratio for MUMM
turbid water atmospheric correction
absaer_opt (int) (default=0) = absorbing aerosol flagging option
0: disabled
1: use rhow_412 aerosol index test
2: GMAO ancillary aerosol test
glint_opt (int) (default=1) = glint correction:
0: glint correction off
1: standard glint correction
2: simple glint correction
oxaband_opt (int) (default=0) = oxygen A-band correction
0: no correction
1: Apply Ding and Gordon (1995) correction
2: Apply oxygen transmittance from gas transmittance table (see gas_opt)
cirrus_opt (boolean) (default=false) = cirrus cloud reflectance correction option
filter_opt (boolean) (default=false) = filtering input data option
filter_file (ifile) (default=$OCDATAROOT/sensor/sensor_filter.dat) =
data file for input filtering
brdf_opt (int) (default=0) = Bidirectional reflectance correction
0: no correction
1: Fresnel reflection/refraction correction for sensor path
3: Fresnel reflection/refraction correction for sensor + solar path
7: Morel f/Q + Fresnel solar + Fresnel sensor
15: Gordon DT + Morel f/Q + Fresnel solar + Fresnel sensor
19: Morel Q + Fresnel solar + Fresnel sensor
fqfile (ifile) (default=$OCDATAROOT/common/morel_fq.nc) = f/Q correction file
parfile (ifile) = par climatology file for NPP calculation
gas_opt (int) (default=1) = gaseous transmittance bitmask selector
0: no correction
1: Ozone
2: CO2
4: NO2
8: H2O
16: Use ATREM
32: Use <sensor>_gas_transmittance.nc tables
64: CO
128: CH4
256: N2O
atrem_opt (int) (default=0) = ATREM gaseous transmittance bitmask selector
0: H2O only
1: Ozone
2: CO2
4: NO2
8: CO
16: CH4
32: O2
64: N2O
atrem_full (int) (default=0) = ATREM gaseous transmittance calculation option
0: Calculate transmittance using k-distribution method (fast)
1: Calculate transmittance using full method (slow)
atrem_geom (int) (default=0) = ATREM gaseous transmittance geometry option
0: Only recalculate geometry when error threshold reached (fast)
1: Recalculate geometry every pixel (slow)
atrem_model (int) (default=0) = ATREM gaseous transmittance Atm. model selection
0: Use pixel's latitude and date to determine model
1: tropical
2: mid latitude summer
3: mid latitude winter
4: subarctic summer
5: subarctic winter
6: US standard 1962
atrem_splitpaths (int) (default=0) = ATREM gaseous transmittance split paths between solar and sensor (turns atrem_full on)
0: Calculates transmittance over total path length (default)
1: Calculates transmittance over separate solar and sensor paths (slow)
iop_opt (int) (default=0) = IOP model for use in downstream products
0: None (products requiring a or bb will fail)
1: Carder
2: GSM
3: QAA
4: PML
5: NIWA
6: LAS
7: GIOP
cphyt_opt (int) (default=1) = model for phytoplankton carbon
1: Graff/Westberry
2: Behrenfeld
seawater_opt (int) (default=0) = seawater IOP options
0: static values
1: temperature & salinity-dependent seawater nw, aw, bbw
polfile (ifile) = polarization sensitivities filename leader
pol_opt (int) (default=-1) = polarization correction (sensor-specific)
0: no correction
1: only Rayleigh component is polarized
2: all radiance polarized like Rayleigh
3: only Rayleigh and Glint are polarized (MODIS default)
4: all radiance polarized like Rayleigh + Glint
band_shift_opt (int) (default=0) = bandshifting option
1: apply bio-optical bandshift
0: linear interpolation
giop_aph_opt (int) (default=2) = GIOP model aph function type
0: tabulated (supplied via giop_aph_file)
2: Bricaud et al. 1995 (chlorophyll supplied via default empirical algorithm)
3: Ciotti and Bricaud 2006 (size fraction supplied via giop_aph_s)
giop_aph_file (ifile) (default=$OCDATAROOT/common/aph_default.txt) =
GIOP model, tabulated aph spectra
giop_uaph_file (ifile) (default=$OCDATAROOT/common/aph_unc_default.txt) =
GIOP model, tabulated aph uncertainty spectra
giop_aph_s (float) (default=-1000.0) = GIOP model, spectral parameter
for aph
giop_adg_opt (int) (default=1) = GIOP model adg function type
0: tabulated (supplied via giop_adg_file)
1: exponential with exponent supplied via giop_adg_s)
2: exponential with exponent derived via Lee et al. (2002)
3: exponential with exponent derived via OBPG method
giop_adg_file (string) (default=$OCDATAROOT/common/adg_default.txt) =
GIOP model, tabulated adg spectra
giop_uadg_file (ifile) (default=$OCDATAROOT/common/adg_unc_default.txt) =
GIOP model, tabulated adg uncertainty spectra
giop_adg_s (float) (default=0.018) = GIOP model, spectral parameter
for adg
giop_uadg_s (float) (default=0.0) = GIOP model, uncertainty of spectral
parameter for adg
giop_bbp_opt (int) (default=3) = GIOP model bbp function type
0: tabulated (supplied via giop_bbp_file)
1: power-law with exponent supplied via giop_bbp_s)
2: power-law with exponent derived via Hoge & Lyon (1996)
3: power-law with exponent derived via Lee et al. (2002)
5: power-law with exponent derived via Ciotti et al. (1999)
6: power-law with exponent derived via Morel & Maritorena (2001)
7: power-law with exponent derived via Loisel & Stramski (2000)
8: spectrally independent vector derived via Loisel & Stramski (2000)
9: fixed vector derived via Loisel & Stramski (2000)
10: fixed vector derived via lee et al. (2002)
11: fixed vector derived via McKinna et al. (2021)
12: fixed vector derived via Huot et al. (2008)
giop_bbp_file (ifile) (default=$OCDATAROOT/common/bbp_default.txt) =
GIOP model, tabulated bbp spectra
giop_ubbp_file (ifile) (default=$OCDATAROOT/common/bbp_default.txt) =
GIOP model, tabulated bbp uncertainty spectra
giop_bbp_s (float) (default=-1000.0) = GIOP model, spectral parameter
for bbp
giop_ubbp_s (float) (default=0.0) = GIOP model, uncertainty in spectral
parameter for bbp
giop_acdom_opt (int) (default=1) = GIOP model acdom function type
0: tabulated (supplied via giop_acdom_file)
1: no data
giop_acdom_file (ifile) =
GIOP model, file of specific CDOM absorption coefficients for aLMI
giop_uacdom_file (ifile) =
GIOP model, file of specific CDOM absorption uncertainties for aLMI
giop_anap_opt (int) (default=1) = GIOP model anap function type
0: tabulated (supplied via giop_anap_file)
1: no data
giop_anap_file (ifile) =
GIOP model, file of specific NAP absorption coefficients for aLMI
giop_uanap_file (ifile) =
GIOP model, file of specific NAP absorption coefficient uncertainties for aLMI
giop_bbph_opt (int) (default=1) = GIOP model bbph function type
0: tabulated (supplied via giop_bbph_file)
1: no data
giop_bbph_file (ifile) =
GIOP model, file of specific phytoplankton backscattering coefficients for aLMI
giop_ubbph_file (ifile) =
GIOP model, file of specific phytoplankton backscatteirng coefficient uncertainties for aLMI
giop_bbnap_opt (int) (default=1) = GIOP model bbnap function type
0: tabulated (supplied via giop_bbnap_file)
1: no data
giop_bbnap_file (ifile) =
GIOP model, file of specific NAP backscattering coefficients for aLMI
giop_ubbnap_file (ifile) =
GIOP model, file of specific NAP backscattering coefficient uncertainties for aLMI
giop_rrs_opt (int) (default=0) = GIOP model Rrs to bb/(a+bb) method
0: Gordon quadratic (specified with giop_grd)
1: Morel f/Q
giop_rrs_unc_opt (int) (default=0) = GIOP model Rrs uncertainties option
0: none
1: sensor-derived Rrs uncertainties
2: input spectral absolute uncertainties using giop_rrs_unc, as comma-separated
array with spectral resolution matching sensor
3: input spectral relative uncertainties using giop_rrs_unc, as comma-separated
array with spectral resolution matching sensor
giop_rrs_diff (float) (default=0.33) = GIOP model, maximum difference between input and modeled Rrs
giop_grd (float) (default=[0.0949,0.0794]) = GIOP model, Gordon
Rrs to bb/(a+bb) quadratic coefficients
giop_wave (float) (default=-1) = GIOP model list of sensor wavelengths for
optimization comma-separated list, default is all visible bands (400-700nm)
giop_maxiter (int) (default=50) = GIOP Model iteration limit
giop_fit_opt (int) (default=1) = GIOP model optimization method
0: Amoeba optimization
1: Levenberg-Marquardt optimization
3: SVD matrix inversion
4: SIOP adaptive matrix inversion
gsm_opt (int) (default=0) = GSM model options
0: default coefficients
1: Chesapeake regional coefficients
gsm_fit (int) (default=0) = GSM fit algorithm
0: Amoeba
1: Levenberg-Marquardt
gsm_adg_s (float) (default=0.02061) = GSM IOP model, spectral slope for adg
gsm_bbp_s (float) (default=1.03373) = GSM IOP model, spectral slope for bbp
gsm_aphw (float) (default=[412.0, 443.0, 490.0, 510.0, 555.0, 670.0]) =
GSM IOP model, wavelengths of ap* table
gsm_aphs (float) (default=[0.00665, 0.05582, 0.02055, 0.01910, 0.01015, 0.01424]) = GSM IOP model, coefficients of ap* table
qaa_adg_s (float) (alias=qaa_S) (default=0.015) = QAA IOP model, spectral
slope for adg
qaa_wave (int) = sensor wavelengths for QAA algorithm
chloc2_wave (int) (default=[-1,-1]) = sensor wavelengths for OC2 chlorophyll
algorithm
chloc2_coef (float) (default=[0.0,0.0,0.0,0.0,0.0]) = coefficients for OC2
chlorophyll algorithm
chloc3_wave (int) (default=[-1,-1,-1]) = sensor wavelengths for OC3
chlorophyll algorithm
chloc3_coef (float) (default=[0.0,0.0,0.0,0.0,0.0]) = coefficients for OC3
chlorophyll algorithm
chloc4_wave (int) (default=[-1,-1,-1,-1]) = sensor wavelengths for OC4
chlorophyll algorithm
chloc4_coef (float) (default=[0.0,0.0,0.0,0.0,0.0]) = coefficients for OC4
chlorophyll algorithm
kd2_wave (int) (default=[-1,-1]) = sensor wavelengths for polynomial Kd(490)
algorithm
kd2_coef (float) (default=[0.0,0.0,0.0,0.0,0.0,0.0]) = sensor wavelengths
for polynomial Kd(490) algorithm
flh_offset (float) (default=0.0) = bias to subtract
from retrieved fluorescence line height
vcnnfile (ifile) = virtual constellation neural net file
picfile (ifile) = pic table for Balch 2-band algorithm
owtfile (ifile) = optical water type file
owtchlerrfile (ifile) = chl error file associate with optical water type
aermodfile (ifile) = aerosol model filename leader
uncertaintyfile (ifile) = uncertainty LUT
aermodels (string) (default=[r30f95v01,r30f80v01,r30f50v01,r30f30v01,r30f20v01,r30f10v01,r30f05v01,r30f02v01,r30f01v01,r30f00v01,r50f95v01,r50f80v01,r50f50v01,r50f30v01,r50f20v01,r50f10v01,r50f05v01,r50f02v01,r50f01v01,r50f00v01,r70f95v01,r70f80v01,r70f50v01,r70f30v01,r70f20v01,r70f10v01,r70f05v01,r70f02v01,r70f01v01,r70f00v01,r75f95v01,r75f80v01,r75f50v01,r75f30v01,r75f20v01,r75f10v01,r75f05v01,r75f02v01,r75f01v01,r75f00v01,r80f95v01,r80f80v01,r80f50v01,r80f30v01,r80f20v01,r80f10v01,r80f05v01,r80f02v01,r80f01v01,r80f00v01,r85f95v01,r85f80v01,r85f50v01,r85f30v01,r85f20v01,r85f10v01,r85f05v01,r85f02v01,r85f01v01,r85f00v01,r90f95v01,r90f80v01,r90f50v01,r90f30v01,r90f20v01,r90f10v01,r90f05v01,r90f02v01,r90f01v01,r90f00v01,r95f95v01,r95f80v01,r95f50v01,r95f30v01,r95f20v01,r95f10v01,r95f05v01,r95f02v01,r95f01v01,r95f00v01]) = aerosol models
met1 (ifile) (default=$OCDATAROOT/common/met_climatology.hdf) =
1st meteorological ancillary data file
met2 (ifile) = 2nd meteorological ancillary data file
met3 (ifile) = 3rd meteorological ancillary data file
ozone1 (ifile) (default=$OCDATAROOT/common/ozone_climatology.hdf) =
1st ozone ancillary data file
ozone2 (ifile) = 2nd ozone ancillary data file
ozone3 (ifile) = 3rd ozone ancillary data file
rad1 (ifile) = Ancillary data for PAR, 1st file
rad2 (ifile) = Ancillary data for PAR, 2nd file
rad3 (ifile) = Ancillary data for PAR, 3rd file
anc_profile1 (ifile) =
1st ancillary profile data file
anc_profile2 (ifile) =
2nd ancillary profile data file
anc_profile3 (ifile) =
3rd ancillary profile data file
anc_aerosol1 (ifile) =
1st ancillary aerosol data file
anc_aerosol2 (ifile) =
2nd ancillary aerosol data file
anc_aerosol3 (ifile) =
3rd ancillary aerosol data file
sfc_albedo (ifile) =
ancillary cloud albedo data file
cth_albedo (ifile) =
ancillary albedo file for cloud top height use
anc_cor_file (ifile) = ozone correction file
pixel_anc_file (ifile) = per pixel ancillary data file
land (ifile) (default=$OCDATAROOT/common/gebco_ocssw_v2020.nc) = land mask file
water (ifile) (default=$OCDATAROOT/common/gebco_ocssw_v2020.nc) =
shallow water mask file
demfile (ifile) (default=$OCDATAROOT/common/gebco_ocssw_v2020.nc) =
digital elevation map file
dem_auxfile (ifile) = auxiliary digital elevation map file
mldfile (ifile) = Multi-layer depth file
icefile (ifile) (default=$OCDATAROOT/common/ice_mask.hdf) = sea ice file
ice_threshold (float) (default=0.1) = sea ice fraction above which will be
flagged as sea ice
sstcoeffile (ifile) = IR sst algorithm coefficients file
dsdicoeffile (ifile) = SST dust correction algorithm coefficients file
sstssesfile (ifile) = IR sst algorithm error statistics file
sst4coeffile (ifile) = SWIR sst algorithm coefficients file
sst4ssesfile (ifile) = SWIR sst algorithm error statistics file
sst3coeffile (ifile) = Triple window sst algorithm coefficients file
sst3ssesfile (ifile) = Triple window sst algorithm error statistics file
sstfile (ifile) (default=$OCDATAROOT/common/sst_climatology.hdf) = input
SST reference file
sstreftype (int) (default=0) = Reference SST field source
0: Reynolds OI SST reference file
1: AMSR-E daily SST reference file
2: AMSR-E 3-day SST reference file
3: ATSR monthly SST reference file
4: NTEV2 monthly SST reference file
5: AMSR-E 3-day or night SST reference file
6: WindSat daily SST reference file
7: WindSat 3-day SST reference file
8: WindSat 3-day or night SST reference file
sstrefdif (float) (default=100.0) = stricter sst-ref difference threshold
viirsnv7 (int) (default=-1) = =1 to use the VIIRSN V7 high senz latband sst and sst3 equations
viirsnosisaf (int) (default=0) = =1 to use the VIIRSN OSI-SAF sst and sst3 equations
no2file (ifile) (default=$OCDATAROOT/common/no2_climatology.hdf) = no2
ancillary file
alphafile (ifile) (default=$OCDATAROOT/common/alpha510_climatology.hdf) =
alpha510 climatology file
tauafile (ifile) (default=$OCDATAROOT/common/taua865_climatology.hdf) =
taua865 climatology file
flaguse (string) (default=ATMFAIL,LAND,HIGLINT,HILT,HISATZEN,STRAYLIGHT,CLDICE,COCCOLITH,LOWLW,CHLFAIL,NAVWARN,ABSAER,MAXAERITER,ATMWARN,HISOLZEN,NAVFAIL) = Flags to use
xcalbox (int) (default=0) = pixel size of the central box in the L1 scene
(e.g. 5 pixels around MOBY) to be extracted into xcalfile for the
cross-calibration, 0=whole L1
xcalboxcenter (int) (default=[0,0]) = Central [ipix, iscan] of the box in
the L1 scene, [0,0] = center of the L1 scene
xcalpervalid (int) (default=0) = min percent of valid cross-calibration
pixels within the box or the L1 scene, 0 = at least 1 pixel
xcalsubsmpl (int) (default=1) = Sub-sampling rate for the data to be used
for the cross-calibration
chlthreshold (float) (default=100.000000) = threshold on L2 data chlorophyll
(100.000000=CHL_MAX)
aotthreshold (float) (default=1.000000) = threshold on L2 data AOTs
(1.000000=AOT_MAX)
coccolith (float) (default=[1.1,0.9,0.75,1.85,1.0,1.65,0.6,1.15]) =
coccolithophore algorithm coefs
cirrus_thresh (float) (default=[-1.0,-1.0]) = cirrus reflectance thresholds
taua (float) = [taua_band1,...,taua_bandn] aerosol optical thickness of the
calibration data point
absaer (float) (default=0.0) = absorbing aerosol threshold on aerosol index
rhoamin (float) (default=0.0001) = min NIR aerosol reflectance to attempt
model lookup
epsmin (float) (default=0.85) = minimum epsilon to trigger atmospheric
correction failure flag
epsmax (float) (default=1.35) = maximum epsilon to trigger atmospheric
correction failure flag
tauamax (float) (default=0.3) = maximum 865 aerosol optical depth to trigger
hitau flag
nLwmin (float) (default=0.15) = minimum nLw(555) to trigger low Lw flag
wsmax (float) (default=12.0) = windspeed limit on white-cap correction in m/s
windspeed (float) (default=-1000.0) = user over-ride of windspeed in m/s
(-1000=use ancillary files)
windangle (float) (default=-1000.0) = user over-ride of wind angle in deg
(-1000=use ancillary files)
pressure (float) (default=-1000.0) = user over-ride of atmospheric pressure
in mb (-1000=use ancillary files)
ozone (float) (default=-1000.0) = user over-ride of ozone concentration in
cm (-1000=use ancillary files)
relhumid (float) (default=-1000.0) = user over-ride of relative humidity in
percent (-1000=use ancillary files)
watervapor (float) (default=-1000.0) = user over-ride of water vapor in
g/cm^2 (-1000=use ancillary files)
vcal_opt (int) (default=-1) = Vicarious calibration option
vcal_chl (float) (default=-1.0) = Vicarious calibration chl
vcal_solz (float) (default=-1.0) = Vicarious calibration solz
vcal_nLw (float) = Vicarious calibration normalized water leaving radiances
vcal_Lw (float) = Vicarious calibration water leaving radiances
vcal_depth (float) (default=-1000.0) = depth to use to exclude data from target file
e.g. -1000 excludes depths less than 1000m
vcal_min_nbin (int) (default=4) = minimum # of samples in a bin for acceptance
vcal_min_nscene (int) (default=3) = minimum # of scenes in a bin for acceptance
owmcfile (ifile) (default=$OCDATAROOT/common/owmc_lut.hdf) = lut for OWMC
classification
north (float) (default=-999) = north boundary
south (float) (default=-999) = south boundary
east (float) (default=-999) = east boundary
west (float) (default=-999) = west boundary
xbox (int) (default=-1) = number of pixels on either side of the SW point
ybox (int) (default=-1) = number of scan lines on either side of the SW point
subsamp (int) (default=1) = sub-sampling interval
prodxmlfile (ofile) = output XML file describing all possible products
breflectfile (ifile) = input NetCDF file for bottom reflectances and bottom types
bpar_validate_opt (int) (default=0) = use solar noon(0), use sensor overpass (1)
bpar_elev_opt (int) (default=0) = use bathymery (0), user-defined geometric depth (1)
bpar_elev_value (float) (default=30.0) = user defined bpar geometric depth value
deflate (int) (default=0) = deflation level
raman_opt (int) (default=0) = Raman scattering Rrs correction options
0: no correction
1: Lee et al. (2013) empirical correction
2: Westberry et al. (2013) analytical correction
3: Lee et al. (1994) analytical correction
gmpfile (ifile) = GMP geometric parameter file (MISR only)
water_spectra_file (ifile) = water absorption/scattering coefficient file
shallow_water_depth (float) (default=30.0) = threshold for flagging shallow water
avw_coef (float) (default=[0.0,0.0,0.0,0.0,0.0,0.0]) = coefficients for AVW
cloud_hgt_file (ifile) = Cloud height and temperature file
Currently, use the MODIS L2 cloud file (MYD06...)
from the LAADS DAAC
doi (string) = Digital Object Identifier (DOI) string
wavelength_3d (string) = wavelength_3d input, written in ascending order
with format 'wavelength_3d=nnn,nnn,nnn' where nnn is a sensor wavelength
or a range of wavelengths as follows 'nnn:nnn'
mbac_wave (int) = bands used for mbac atmospheric correction
calfile (ifile) = system calibration file
rad_opt (int) (default=0) = radiation correction option (sensor-specific)
0: no correction
1: apply MERIS Smile correction
viirscalparfile (ifile) = VIIRS L1A calibration parameter file name (VIIRS only)
geom_per_band (boolean) (default=0) = geometry per band option:
0: use nominal viewing geometry - same for all bands
1: use band-specific viewing geometry (if available)
xcalfile (ifile) = cross-calibration file
xcal_opt (int) = cross-calibration option (sensor-specific) comma separated
list of option values, 1 per band, with bands listed in xcal_wave.
3: apply cross-calibration corrections (polarization and rvs)
2: apply cross-calibration polarization corrections
1: apply cross-calibration rvs corrections
0: no correction
xcal_wave (float) = wavelengths at which to apply cross-calibration. Comma
separated list of sensor wavelength values associated with xcal_opt.
resolution (int) (default=-1) = processing resolution (MODIS only)
-1: standard ocean 1km processing
1000: 1km resolution including aggregated 250 and 500m land bands
500: 500m resolution including aggregated 250 land bands and
replication for lower resolution bands
250: 250m resolution with replication for lower resolution bands
newavhrrcal (int) (default=0) = =1 for new noaa-16 calibration
ch22detcor (float) (default=[1.0,1.0,1.0,1.0,1.0,1.0,1.0,1.0,1.0,1.0]) =
Channel 22 detector corrections (MODIS only)
ch23detcor (float) (default=[1.0,1.0,1.0,1.0,1.0,1.0,1.0,1.0,1.0,1.0]) =
Channel 23 detector corrections (MODIS only)
sl_pixl (int) (default=-1) = SeaWiFS only, number of LAC pixels for
straylight flagging
sl_frac (float) (default=0.25) = SeaWiFS only, straylight fractional
threshold on Ltypical
outband_opt (int) (default=99) = out-of-band correction for water-leaving
radiances
2: On (default for MODIS, SeaWiFS, OCTS)
0: Off (default for MOS, OSMI)
maskland (boolean) (default=on) = land mask option
maskbath (boolean) (default=off) = shallow water mask option
maskcloud (boolean) (default=on) = cloud mask option
maskglint (boolean) (default=off) = glint mask option
masksunzen (boolean) (default=off) = large sun zenith angle mask option
masksatzen (boolean) (default=off) = large satellite zenith angle mask option
maskhilt (boolean) (default=on) = high Lt masking
maskstlight (boolean) (default=on) = stray light masking
sunzen (float) (default=75.0) = sun zenith angle threshold in deg.
satzen (float) (default=60.0) = satellite zenith angle threshold
hipol (float) (default=0.5) = threshold on degree-of-polarization to set
HIPOL flag
glint_thresh (float) (alias=glint) (default=0.005) = high sun glint threshold
cloud_thresh (float) (alias=albedo) (default=0.027) = cloud reflectance
threshold
cloud_wave (float) (default=865.0) = wavelength of cloud reflectance test
cloud_eps (float) (default=-1.0) = cloud reflectance ratio threshold
(-1.0=disabled)
cloud_mask_file (ifile) = cloud mask file
cloud_mask_opt (int) (default=0) = cloud mask file variable name
0: cloud_flag
1: cloud_flag_dilated
offset (float) = calibration offset adjustment
gain (float) = calibration gain multiplier
spixl (int) (default=1) = start pixel number
epixl (int) (default=-1) = end pixel number (-1=the last pixel)
dpixl (int) (default=1) = pixel sub-sampling interval
sline (int) (default=1) = start line number
eline (int) (default=-1) = end line number (-1=the last line)
dline (int) (default=1) = line sub-sampling interval
georegion_file (string) = geo region mask file with lat,lon,georegion 1=process pixel, 0=do not
To process a L1B file using l2gen, at a minimum, you need to set an infile name (ifile) and an outfile name (ofile). You can also indicate a data suite; in this example, we will proceed with the biogeochemical (BGC) suite that includes chlorophyll a estimates.
Parameters can be passed to OCSSW programs through a text file. They can also be passed as arguments, but writing to a text file leaves a clear processing record. Define the parameters in a dictionary, then send it to the write_par function
defined in the Setup section.
par = {
"ifile": l2gen_ifile,
"ofile": str(l2gen_ifile).replace(".L1B.", ".L2_SFREFL."),
"suite": "SFREFL",
"atmocor": 0,
}
write_par("l2gen.par", par)
With the parameter file ready, it’s time to call l2gen from a %%bash cell.
%%bash
source $OCSSWROOT/OCSSW_bash.env
l2gen par=l2gen.par
Loading default parameters from /opt/ocssw/share/common/msl12_defaults.par
Input file data/PACE_OCI.20240605T092137.L1B.V2.nc is PACE L1B file.
Loading characteristics for OCI
Opening sensor information file /opt/ocssw/share/oci/msl12_sensor_info.dat
Bnd Lam Fo Tau_r k_oz k_no2 t_co2 awhite aw bbw
0 314.550 112.026 4.873e-01 4.208e-01 3.281e-19 1.000e+00 0.000e+00 2.305e-01 6.356e-03
1 316.239 92.478 6.485e-01 5.806e-01 2.961e-19 1.000e+00 0.000e+00 1.633e-01 7.727e-03
2 318.262 85.195 7.410e-01 5.473e-01 2.844e-19 1.000e+00 0.000e+00 1.278e-01 8.187e-03
3 320.303 82.175 7.809e-01 4.609e-01 2.833e-19 1.000e+00 0.000e+00 1.105e-01 8.271e-03
4 322.433 80.733 7.906e-01 3.543e-01 2.898e-19 1.000e+00 0.000e+00 9.950e-02 8.190e-03
5 324.649 86.251 7.915e-01 2.567e-01 3.018e-19 1.000e+00 0.000e+00 9.079e-02 8.041e-03
6 326.828 95.932 7.891e-01 1.907e-01 3.132e-19 1.000e+00 0.000e+00 8.475e-02 7.871e-03
7 328.988 101.672 7.700e-01 1.386e-01 3.251e-19 1.000e+00 0.000e+00 8.211e-02 7.627e-03
8 331.305 101.708 7.404e-01 9.852e-02 3.417e-19 1.000e+00 0.000e+00 8.089e-02 7.342e-03
9 333.958 97.745 7.204e-01 6.830e-02 3.572e-19 1.000e+00 0.000e+00 7.656e-02 7.132e-03
10 336.815 93.497 7.112e-01 4.395e-02 3.736e-19 1.000e+00 0.000e+00 6.891e-02 6.975e-03
11 339.160 96.099 7.034e-01 3.070e-02 3.963e-19 1.000e+00 0.000e+00 6.288e-02 6.834e-03
12 341.321 98.332 6.898e-01 2.060e-02 4.076e-19 1.000e+00 0.000e+00 5.915e-02 6.663e-03
13 343.632 95.167 6.720e-01 1.439e-02 4.137e-19 1.000e+00 0.000e+00 5.624e-02 6.474e-03
14 346.017 92.958 6.529e-01 9.882e-03 4.395e-19 1.000e+00 0.000e+00 5.365e-02 6.285e-03
15 348.468 95.422 6.342e-01 6.032e-03 4.610e-19 1.000e+00 0.000e+00 5.106e-02 6.097e-03
16 350.912 99.423 6.163e-01 4.542e-03 4.617e-19 1.000e+00 0.000e+00 4.864e-02 5.913e-03
17 353.344 102.927 5.988e-01 3.473e-03 4.768e-19 1.000e+00 0.000e+00 4.647e-02 5.735e-03
18 355.782 99.180 5.823e-01 2.499e-03 5.019e-19 1.000e+00 0.000e+00 4.450e-02 5.562e-03
19 358.235 93.459 5.642e-01 2.283e-03 5.089e-19 1.000e+00 0.000e+00 4.256e-02 5.394e-03
20 360.695 97.255 5.472e-01 2.096e-03 5.117e-19 1.000e+00 0.000e+00 4.053e-02 5.233e-03
21 363.137 105.330 5.320e-01 1.734e-03 5.326e-19 1.000e+00 0.000e+00 3.856e-02 5.079e-03
22 365.610 113.914 5.178e-01 1.415e-03 5.533e-19 1.000e+00 0.000e+00 3.642e-02 4.932e-03
23 368.083 118.845 5.043e-01 1.282e-03 5.555e-19 1.000e+00 0.000e+00 3.417e-02 4.789e-03
24 370.534 115.549 4.908e-01 1.286e-03 5.590e-19 1.000e+00 0.000e+00 3.126e-02 4.651e-03
25 372.991 107.218 4.773e-01 1.342e-03 5.744e-19 1.000e+00 0.000e+00 2.719e-02 4.520e-03
26 375.482 108.880 4.630e-01 1.283e-03 5.834e-19 1.000e+00 0.000e+00 2.270e-02 4.392e-03
27 377.926 118.137 4.513e-01 1.115e-03 5.861e-19 1.000e+00 0.000e+00 1.843e-02 4.269e-03
28 380.419 111.060 4.406e-01 1.167e-03 5.945e-19 1.000e+00 0.000e+00 1.508e-02 4.151e-03
29 382.876 96.370 4.280e-01 1.379e-03 6.014e-19 1.000e+00 0.000e+00 1.316e-02 4.037e-03
30 385.359 96.688 4.156e-01 1.389e-03 6.039e-19 1.000e+00 0.000e+00 1.228e-02 3.928e-03
31 387.811 107.161 4.051e-01 1.257e-03 6.130e-19 1.000e+00 0.000e+00 1.166e-02 3.822e-03
32 390.297 112.501 3.955e-01 1.196e-03 6.227e-19 1.000e+00 0.000e+00 1.104e-02 3.718e-03
33 392.764 106.686 3.857e-01 1.256e-03 6.193e-19 1.000e+00 0.000e+00 1.065e-02 3.619e-03
34 395.238 106.058 3.747e-01 1.271e-03 6.157e-19 1.000e+00 0.000e+00 1.040e-02 3.523e-03
35 397.706 127.304 3.647e-01 1.082e-03 6.328e-19 1.000e+00 0.000e+00 9.891e-03 3.430e-03
36 400.178 157.144 3.564e-01 9.418e-04 6.342e-19 1.000e+00 0.000e+00 8.906e-03 3.341e-03
37 402.654 170.763 3.486e-01 9.296e-04 6.155e-19 1.000e+00 0.000e+00 7.976e-03 3.255e-03
38 405.127 168.889 3.400e-01 9.759e-04 6.015e-19 1.000e+00 0.000e+00 7.534e-03 3.171e-03
39 407.605 168.988 3.314e-01 1.065e-03 6.101e-19 1.000e+00 0.000e+00 7.203e-03 3.090e-03
40 410.074 173.399 3.233e-01 1.143e-03 6.226e-19 1.000e+00 0.000e+00 6.868e-03 3.011e-03
41 412.557 177.268 3.155e-01 1.158e-03 6.161e-19 1.000e+00 0.000e+00 6.656e-03 2.935e-03
42 415.025 177.759 3.079e-01 1.218e-03 5.856e-19 1.000e+00 0.000e+00 6.564e-03 2.861e-03
43 417.512 175.925 3.004e-01 1.325e-03 5.698e-19 1.000e+00 0.000e+00 6.661e-03 2.789e-03
44 419.988 175.156 2.932e-01 1.448e-03 5.927e-19 1.000e+00 0.000e+00 6.771e-03 2.720e-03
45 422.453 174.012 2.863e-01 1.774e-03 6.054e-19 1.000e+00 0.000e+00 6.878e-03 2.654e-03
46 424.940 170.766 2.796e-01 2.153e-03 5.793e-19 1.000e+00 0.000e+00 7.006e-03 2.590e-03
47 427.398 162.027 2.732e-01 2.242e-03 5.468e-19 1.000e+00 0.000e+00 6.986e-03 2.527e-03
48 429.885 154.088 2.664e-01 2.290e-03 5.331e-19 1.000e+00 0.000e+00 6.994e-03 2.464e-03
49 432.379 161.564 2.597e-01 2.492e-03 5.452e-19 1.000e+00 0.000e+00 7.152e-03 2.404e-03
50 434.869 174.172 2.539e-01 2.644e-03 5.549e-19 1.000e+00 0.000e+00 7.438e-03 2.347e-03
51 437.351 178.115 2.482e-01 3.023e-03 5.355e-19 1.000e+00 0.000e+00 8.014e-03 2.291e-03
52 439.828 182.229 2.424e-01 3.903e-03 4.968e-19 1.000e+00 0.000e+00 8.606e-03 2.238e-03
53 442.327 190.795 2.368e-01 4.600e-03 4.726e-19 1.000e+00 0.000e+00 9.139e-03 2.185e-03
54 444.811 195.628 2.315e-01 4.590e-03 4.945e-19 1.000e+00 0.000e+00 9.759e-03 2.133e-03
55 447.309 199.848 2.261e-01 4.547e-03 5.107e-19 1.000e+00 0.000e+00 1.052e-02 2.082e-03
56 449.795 205.764 2.211e-01 4.968e-03 4.798e-19 1.000e+00 0.000e+00 1.131e-02 2.034e-03
57 452.280 205.708 2.162e-01 5.368e-03 4.441e-19 1.000e+00 0.000e+00 1.179e-02 1.987e-03
58 454.769 204.450 2.114e-01 5.945e-03 4.252e-19 1.000e+00 0.000e+00 1.203e-02 1.942e-03
59 457.262 205.953 2.067e-01 7.420e-03 4.263e-19 1.000e+00 0.000e+00 1.212e-02 1.898e-03
60 459.757 207.117 2.022e-01 9.301e-03 4.351e-19 1.000e+00 0.000e+00 1.214e-02 1.855e-03
61 462.252 207.870 1.977e-01 1.011e-02 4.326e-19 1.000e+00 0.000e+00 1.219e-02 1.813e-03
62 464.737 205.886 1.935e-01 9.683e-03 4.061e-19 1.000e+00 0.000e+00 1.238e-02 1.772e-03
63 467.244 203.675 1.893e-01 9.647e-03 3.691e-19 1.000e+00 0.000e+00 1.242e-02 1.733e-03
64 469.729 203.713 1.852e-01 1.043e-02 3.441e-19 1.000e+00 0.000e+00 1.244e-02 1.694e-03
65 472.202 205.281 1.812e-01 1.115e-02 3.582e-19 1.000e+00 0.000e+00 1.276e-02 1.657e-03
66 474.700 207.058 1.774e-01 1.242e-02 3.801e-19 1.000e+00 0.000e+00 1.326e-02 1.621e-03
67 477.189 208.385 1.736e-01 1.528e-02 3.580e-19 1.000e+00 0.000e+00 1.393e-02 1.586e-03
68 479.689 209.553 1.700e-01 1.895e-02 3.338e-19 1.000e+00 0.000e+00 1.437e-02 1.552e-03
69 482.183 206.590 1.665e-01 2.122e-02 3.022e-19 1.000e+00 0.000e+00 1.472e-02 1.518e-03
70 484.689 195.340 1.631e-01 2.114e-02 2.725e-19 1.000e+00 0.000e+00 1.533e-02 1.485e-03
71 487.182 189.524 1.595e-01 2.040e-02 2.939e-19 1.000e+00 0.000e+00 1.612e-02 1.454e-03
72 489.674 195.470 1.562e-01 2.101e-02 3.044e-19 1.000e+00 0.000e+00 1.694e-02 1.423e-03
73 492.176 199.211 1.530e-01 2.224e-02 2.853e-19 1.000e+00 0.000e+00 1.789e-02 1.393e-03
74 494.686 200.274 1.499e-01 2.354e-02 2.822e-19 1.000e+00 0.000e+00 1.916e-02 1.364e-03
75 497.182 199.355 1.469e-01 2.613e-02 2.498e-19 1.000e+00 0.000e+00 2.072e-02 1.336e-03
76 499.688 194.720 1.440e-01 3.087e-02 2.022e-19 1.000e+00 0.000e+00 2.233e-02 1.309e-03
77 502.190 192.753 1.410e-01 3.665e-02 2.151e-19 1.000e+00 0.000e+00 2.438e-02 1.281e-03
78 504.695 195.557 1.382e-01 4.082e-02 2.384e-19 1.000e+00 0.000e+00 2.700e-02 1.255e-03
79 507.198 197.605 1.355e-01 4.179e-02 2.315e-19 1.000e+00 0.000e+00 3.003e-02 1.230e-03
80 509.720 196.520 1.328e-01 4.101e-02 2.296e-19 1.000e+00 0.000e+00 3.389e-02 1.206e-03
81 512.213 193.098 1.302e-01 4.110e-02 2.157e-19 1.000e+00 0.000e+00 3.784e-02 1.181e-03
82 514.729 185.769 1.277e-01 4.275e-02 1.772e-19 1.000e+00 0.000e+00 4.045e-02 1.157e-03
83 517.219 179.326 1.252e-01 4.504e-02 1.597e-19 1.000e+00 0.000e+00 4.179e-02 1.134e-03
84 519.747 182.521 1.227e-01 4.767e-02 1.620e-19 1.000e+00 0.000e+00 4.270e-02 1.111e-03
85 522.249 189.089 1.203e-01 5.134e-02 1.624e-19 1.000e+00 0.000e+00 4.337e-02 1.089e-03
86 524.771 188.823 1.180e-01 5.654e-02 1.755e-19 1.000e+00 0.000e+00 4.387e-02 1.067e-03
87 527.276 187.920 1.158e-01 6.292e-02 1.771e-19 1.000e+00 0.000e+00 4.454e-02 1.049e-03
88 529.798 191.821 1.137e-01 6.884e-02 1.592e-19 1.000e+00 0.000e+00 4.553e-02 1.031e-03
89 532.314 193.286 1.114e-01 7.264e-02 1.423e-19 1.000e+00 0.000e+00 4.646e-02 1.008e-03
90 534.859 191.915 1.092e-01 7.422e-02 1.168e-19 1.000e+00 0.000e+00 4.742e-02 9.861e-04
91 537.346 190.229 1.072e-01 7.488e-02 1.024e-19 1.000e+00 0.000e+00 4.852e-02 9.673e-04
92 539.878 187.754 1.052e-01 7.665e-02 1.097e-19 1.000e+00 0.000e+00 4.966e-02 9.489e-04
93 542.395 188.298 1.032e-01 7.989e-02 1.240e-19 1.000e+00 0.000e+00 5.116e-02 9.303e-04
94 544.904 189.892 1.013e-01 8.325e-02 1.296e-19 1.000e+00 0.000e+00 5.328e-02 9.125e-04
95 547.441 189.629 9.940e-02 8.591e-02 1.237e-19 1.000e+00 0.000e+00 5.584e-02 8.953e-04
96 549.994 189.342 9.756e-02 8.809e-02 1.129e-19 1.000e+00 0.000e+00 5.851e-02 8.784e-04
97 552.511 189.725 9.575e-02 9.044e-02 9.907e-20 1.000e+00 0.000e+00 6.062e-02 8.617e-04
98 555.044 188.862 9.400e-02 9.359e-02 8.268e-20 1.000e+00 0.000e+00 6.191e-02 8.454e-04
99 557.576 185.867 9.230e-02 9.815e-02 6.751e-20 1.000e+00 0.000e+00 6.298e-02 8.297e-04
100 560.104 184.284 9.062e-02 1.041e-01 6.704e-20 1.000e+00 0.000e+00 6.443e-02 8.146e-04
101 562.642 184.903 8.899e-02 1.101e-01 8.271e-20 1.000e+00 0.000e+00 6.592e-02 7.997e-04
102 565.190 184.929 8.737e-02 1.152e-01 8.817e-20 1.000e+00 0.000e+00 6.740e-02 7.846e-04
103 567.710 184.394 8.579e-02 1.194e-01 8.097e-20 1.000e+00 0.000e+00 6.954e-02 7.698e-04
104 570.259 184.337 8.424e-02 1.231e-01 8.258e-20 1.000e+00 0.000e+00 7.243e-02 7.555e-04
105 572.796 185.701 8.273e-02 1.258e-01 7.445e-20 1.000e+00 0.000e+00 7.601e-02 7.418e-04
106 575.343 185.860 8.130e-02 1.262e-01 5.358e-20 1.000e+00 0.000e+00 8.077e-02 7.290e-04
107 577.902 184.295 7.992e-02 1.245e-01 4.606e-20 1.000e+00 0.000e+00 8.675e-02 7.169e-04
108 580.450 184.370 7.850e-02 1.217e-01 4.637e-20 1.000e+00 0.000e+00 9.411e-02 7.037e-04
109 582.996 184.516 7.711e-02 1.191e-01 4.563e-20 1.000e+00 0.000e+00 1.036e-01 6.904e-04
110 585.553 182.261 7.581e-02 1.175e-01 4.880e-20 1.000e+00 0.000e+00 1.147e-01 6.784e-04
111 588.086 178.467 7.455e-02 1.172e-01 5.159e-20 1.000e+00 0.000e+00 1.271e-01 6.670e-04
112 590.548 177.770 7.336e-02 1.186e-01 5.765e-20 1.000e+00 0.000e+00 1.411e-01 6.570e-04
113 593.084 179.810 7.228e-02 1.222e-01 5.611e-20 1.000e+00 0.000e+00 1.572e-01 6.487e-04
114 595.679 179.992 7.117e-02 1.275e-01 4.249e-20 1.000e+00 0.000e+00 1.772e-01 6.390e-04
115 598.262 178.361 6.999e-02 1.328e-01 3.893e-20 1.000e+00 0.000e+00 2.030e-01 6.276e-04
116 600.545 176.370 6.757e-02 1.365e-01 3.874e-20 1.000e+00 0.000e+00 2.467e-01 5.992e-04
117 602.920 176.174 6.651e-02 1.374e-01 3.034e-20 1.000e+00 0.000e+00 2.611e-01 5.899e-04
118 605.461 176.473 6.543e-02 1.353e-01 2.076e-20 1.000e+00 0.000e+00 2.702e-01 5.801e-04
119 607.986 174.705 6.437e-02 1.310e-01 1.990e-20 1.000e+00 0.000e+00 2.728e-01 5.706e-04
120 610.360 172.381 6.335e-02 1.256e-01 2.700e-20 1.000e+00 0.000e+00 2.733e-01 5.614e-04
121 612.730 170.269 6.236e-02 1.202e-01 3.487e-20 1.000e+00 0.000e+00 2.746e-01 5.524e-04
122 615.145 167.881 6.137e-02 1.152e-01 3.541e-20 1.000e+00 0.000e+00 2.763e-01 5.434e-04
123 617.605 167.707 6.038e-02 1.107e-01 3.180e-20 1.000e+00 0.000e+00 2.790e-01 5.346e-04
124 620.061 168.940 5.942e-02 1.070e-01 2.798e-20 1.000e+00 0.000e+00 2.832e-01 5.259e-04
125 622.530 167.529 5.848e-02 1.038e-01 2.439e-20 1.000e+00 0.000e+00 2.876e-01 5.173e-04
126 624.988 165.875 5.754e-02 1.007e-01 2.069e-20 1.000e+00 0.000e+00 2.918e-01 5.089e-04
127 627.434 166.459 5.663e-02 9.740e-02 1.634e-20 1.000e+00 0.000e+00 2.963e-01 5.006e-04
128 629.898 165.893 5.575e-02 9.400e-02 1.319e-20 1.000e+00 0.000e+00 3.008e-01 4.925e-04
129 632.376 164.212 5.486e-02 9.046e-02 1.304e-20 1.000e+00 0.000e+00 3.055e-01 4.846e-04
130 634.830 163.727 5.400e-02 8.676e-02 1.402e-20 1.000e+00 0.000e+00 3.097e-01 4.768e-04
131 637.305 163.541 5.316e-02 8.287e-02 1.464e-20 1.000e+00 0.000e+00 3.137e-01 4.692e-04
132 639.791 162.086 5.234e-02 7.890e-02 1.583e-20 1.000e+00 0.000e+00 3.191e-01 4.618e-04
133 641.029 161.259 5.193e-02 7.696e-02 1.681e-20 1.000e+00 0.000e+00 3.224e-01 4.581e-04
134 642.255 160.668 5.152e-02 7.512e-02 1.767e-20 1.000e+00 0.000e+00 3.258e-01 4.545e-04
135 643.479 160.372 5.112e-02 7.339e-02 1.849e-20 1.000e+00 0.000e+00 3.293e-01 4.509e-04
136 644.716 160.231 5.073e-02 7.176e-02 1.919e-20 1.000e+00 0.000e+00 3.327e-01 4.473e-04
137 645.966 159.889 5.034e-02 7.021e-02 1.946e-20 1.000e+00 0.000e+00 3.360e-01 4.437e-04
138 647.188 159.260 4.995e-02 6.871e-02 1.898e-20 1.000e+00 0.000e+00 3.396e-01 4.402e-04
139 648.435 158.731 4.956e-02 6.727e-02 1.756e-20 1.000e+00 0.000e+00 3.436e-01 4.367e-04
140 649.667 158.341 4.918e-02 6.588e-02 1.544e-20 1.000e+00 0.000e+00 3.486e-01 4.333e-04
141 650.913 158.275 4.880e-02 6.453e-02 1.328e-20 1.000e+00 0.000e+00 3.546e-01 4.299e-04
142 652.153 157.789 4.843e-02 6.321e-02 1.167e-20 1.000e+00 0.000e+00 3.616e-01 4.265e-04
143 653.388 155.177 4.807e-02 6.193e-02 1.068e-20 1.000e+00 0.000e+00 3.693e-01 4.231e-04
144 654.622 151.698 4.771e-02 6.060e-02 9.722e-21 1.000e+00 0.000e+00 3.774e-01 4.198e-04
145 655.869 148.463 4.733e-02 5.922e-02 8.805e-21 1.000e+00 0.000e+00 3.858e-01 4.166e-04
146 657.101 147.695 4.695e-02 5.782e-02 7.987e-21 1.000e+00 0.000e+00 3.947e-01 4.133e-04
147 658.340 149.658 4.658e-02 5.648e-02 7.170e-21 1.000e+00 0.000e+00 4.036e-01 4.101e-04
148 659.600 152.493 4.623e-02 5.523e-02 6.329e-21 1.000e+00 0.000e+00 4.119e-01 4.070e-04
149 660.833 154.868 4.590e-02 5.404e-02 5.541e-21 1.000e+00 0.000e+00 4.192e-01 4.038e-04
150 662.067 155.409 4.555e-02 5.283e-02 4.976e-21 1.000e+00 0.000e+00 4.251e-01 4.007e-04
151 663.300 155.290 4.521e-02 5.158e-02 4.897e-21 1.000e+00 0.000e+00 4.297e-01 3.976e-04
152 664.564 155.005 4.487e-02 5.029e-02 5.478e-21 1.000e+00 0.000e+00 4.334e-01 3.945e-04
153 665.795 154.711 4.453e-02 4.895e-02 6.640e-21 1.000e+00 0.000e+00 4.363e-01 3.915e-04
154 667.023 154.401 4.420e-02 4.759e-02 7.980e-21 1.000e+00 0.000e+00 4.389e-01 3.885e-04
155 668.263 154.028 4.387e-02 4.622e-02 9.022e-21 1.000e+00 0.000e+00 4.415e-01 3.855e-04
156 669.518 153.439 4.354e-02 4.488e-02 9.638e-21 1.000e+00 0.000e+00 4.442e-01 3.825e-04
157 670.755 152.834 4.321e-02 4.356e-02 9.943e-21 1.000e+00 0.000e+00 4.471e-01 3.796e-04
158 671.990 152.273 4.289e-02 4.229e-02 1.010e-20 1.000e+00 0.000e+00 4.497e-01 3.767e-04
159 673.245 151.870 4.257e-02 4.107e-02 1.027e-20 1.000e+00 0.000e+00 4.523e-01 3.738e-04
160 674.503 151.619 4.225e-02 3.991e-02 1.027e-20 1.000e+00 0.000e+00 4.551e-01 3.710e-04
161 675.731 151.421 4.194e-02 3.879e-02 9.926e-21 1.000e+00 0.000e+00 4.583e-01 3.681e-04
162 676.963 151.155 4.163e-02 3.774e-02 9.232e-21 1.000e+00 0.000e+00 4.620e-01 3.654e-04
163 678.208 150.791 4.132e-02 3.675e-02 8.304e-21 1.000e+00 0.000e+00 4.661e-01 3.626e-04
164 679.448 150.430 4.102e-02 3.584e-02 7.379e-21 1.000e+00 0.000e+00 4.703e-01 3.599e-04
165 680.680 149.905 4.072e-02 3.503e-02 6.552e-21 1.000e+00 0.000e+00 4.747e-01 3.572e-04
166 681.919 149.251 4.042e-02 3.430e-02 5.895e-21 1.000e+00 0.000e+00 4.794e-01 3.545e-04
167 683.171 148.503 4.012e-02 3.361e-02 5.490e-21 1.000e+00 0.000e+00 4.845e-01 3.518e-04
168 684.417 147.884 3.983e-02 3.292e-02 5.184e-21 1.000e+00 0.000e+00 4.901e-01 3.492e-04
169 685.657 147.515 3.953e-02 3.216e-02 4.905e-21 1.000e+00 0.000e+00 4.962e-01 3.466e-04
170 686.894 147.425 3.924e-02 3.131e-02 4.525e-21 1.000e+00 0.000e+00 5.029e-01 3.440e-04
171 688.143 147.400 3.896e-02 3.039e-02 4.024e-21 1.000e+00 0.000e+00 5.103e-01 3.414e-04
172 689.394 147.153 3.867e-02 2.943e-02 3.539e-21 1.000e+00 0.000e+00 5.186e-01 3.389e-04
173 690.647 146.715 3.839e-02 2.845e-02 3.144e-21 1.000e+00 0.000e+00 5.276e-01 3.363e-04
174 691.888 146.143 3.811e-02 2.750e-02 2.906e-21 1.000e+00 0.000e+00 5.375e-01 3.338e-04
175 693.130 145.684 3.784e-02 2.657e-02 2.763e-21 1.000e+00 0.000e+00 5.484e-01 3.314e-04
176 694.382 145.196 3.756e-02 2.567e-02 2.656e-21 1.000e+00 0.000e+00 5.605e-01 3.289e-04
177 695.644 144.701 3.729e-02 2.479e-02 2.542e-21 1.000e+00 0.000e+00 5.740e-01 3.265e-04
178 696.891 144.099 3.702e-02 2.394e-02 2.429e-21 1.000e+00 0.000e+00 5.888e-01 3.241e-04
179 698.118 143.280 3.676e-02 2.315e-02 2.343e-21 1.000e+00 0.000e+00 6.047e-01 3.217e-04
180 699.376 142.397 3.649e-02 2.241e-02 2.369e-21 1.000e+00 0.000e+00 6.215e-01 3.193e-04
181 700.612 141.532 3.623e-02 2.174e-02 2.581e-21 1.000e+00 0.000e+00 6.393e-01 3.170e-04
182 701.858 141.039 3.597e-02 2.114e-02 3.030e-21 1.000e+00 0.000e+00 6.580e-01 3.147e-04
183 703.097 140.869 3.571e-02 2.061e-02 3.660e-21 1.000e+00 0.000e+00 6.780e-01 3.124e-04
184 704.354 140.969 3.546e-02 2.014e-02 4.374e-21 1.000e+00 0.000e+00 6.995e-01 3.101e-04
185 705.593 140.996 3.521e-02 1.972e-02 4.920e-21 1.000e+00 0.000e+00 7.231e-01 3.078e-04
186 706.833 140.760 3.496e-02 1.934e-02 5.156e-21 1.000e+00 0.000e+00 7.499e-01 3.056e-04
187 708.089 140.307 3.471e-02 1.900e-02 5.111e-21 1.000e+00 0.000e+00 7.805e-01 3.034e-04
188 709.337 139.705 3.446e-02 1.868e-02 4.920e-21 1.000e+00 0.000e+00 8.152e-01 3.012e-04
189 710.581 139.142 3.422e-02 1.842e-02 4.776e-21 1.000e+00 0.000e+00 8.540e-01 2.990e-04
190 711.826 138.503 3.398e-02 1.820e-02 4.601e-21 1.000e+00 0.000e+00 8.959e-01 2.968e-04
191 713.068 137.918 3.374e-02 1.804e-02 4.294e-21 1.000e+00 0.000e+00 9.408e-01 2.947e-04
192 714.316 137.373 3.350e-02 1.788e-02 3.751e-21 1.000e+00 0.000e+00 9.886e-01 2.926e-04
193 716.817 136.139 3.303e-02 1.733e-02 2.447e-21 1.000e+00 0.000e+00 1.093e+00 2.884e-04
194 719.298 134.747 3.257e-02 1.625e-02 1.823e-21 1.000e+00 0.000e+00 1.206e+00 2.843e-04
195 721.800 134.248 3.212e-02 1.496e-02 1.858e-21 1.000e+00 0.000e+00 1.327e+00 2.802e-04
196 724.303 134.303 3.167e-02 1.391e-02 1.678e-21 1.000e+00 0.000e+00 1.461e+00 2.763e-04
197 726.796 133.424 3.124e-02 1.304e-02 1.167e-21 1.000e+00 0.000e+00 1.642e+00 2.724e-04
198 729.299 132.173 3.081e-02 1.220e-02 8.984e-22 1.000e+00 0.000e+00 1.891e+00 2.686e-04
199 731.790 131.770 3.038e-02 1.153e-02 9.392e-22 1.000e+00 0.000e+00 2.176e+00 2.648e-04
200 734.281 131.420 2.997e-02 1.114e-02 1.157e-21 1.000e+00 0.000e+00 2.438e+00 2.611e-04
201 736.791 130.150 2.956e-02 1.096e-02 1.301e-21 1.000e+00 0.000e+00 2.621e+00 2.575e-04
202 739.287 128.259 2.916e-02 1.090e-02 1.303e-21 1.000e+00 0.000e+00 2.732e+00 2.539e-04
203 740.535 127.659 2.897e-02 1.094e-02 1.339e-21 1.000e+00 0.000e+00 2.770e+00 2.521e-04
204 741.785 127.339 2.877e-02 1.103e-02 1.496e-21 1.000e+00 0.000e+00 2.799e+00 2.504e-04
205 743.046 127.458 2.857e-02 1.119e-02 1.813e-21 1.000e+00 0.000e+00 2.819e+00 2.486e-04
206 744.286 127.660 2.838e-02 1.137e-02 2.155e-21 1.000e+00 0.000e+00 2.832e+00 2.469e-04
207 745.534 127.796 2.819e-02 1.153e-02 2.323e-21 1.000e+00 0.000e+00 2.840e+00 2.452e-04
208 746.789 127.697 2.800e-02 1.159e-02 2.205e-21 1.000e+00 0.000e+00 2.846e+00 2.435e-04
209 748.041 127.310 2.781e-02 1.150e-02 1.876e-21 1.000e+00 0.000e+00 2.850e+00 2.419e-04
210 749.279 126.838 2.762e-02 1.122e-02 1.554e-21 1.000e+00 0.000e+00 2.854e+00 2.402e-04
211 750.540 126.351 2.744e-02 1.074e-02 1.390e-21 1.000e+00 0.000e+00 2.857e+00 2.386e-04
212 751.792 126.028 2.725e-02 1.013e-02 1.397e-21 1.000e+00 0.000e+00 2.862e+00 2.369e-04
213 753.042 125.814 2.707e-02 9.483e-03 1.443e-21 1.000e+00 0.000e+00 2.867e+00 2.353e-04
214 754.294 125.571 2.689e-02 8.866e-03 1.420e-21 1.000e+00 0.000e+00 2.870e+00 2.337e-04
215 755.542 125.152 2.671e-02 8.328e-03 1.307e-21 1.000e+00 0.000e+00 2.872e+00 2.321e-04
216 756.802 124.719 2.654e-02 7.888e-03 1.121e-21 1.000e+00 0.000e+00 2.871e+00 2.305e-04
217 758.051 124.328 2.636e-02 7.536e-03 8.963e-22 1.000e+00 0.000e+00 2.869e+00 2.290e-04
218 759.299 124.043 2.618e-02 7.263e-03 6.951e-22 1.000e+00 0.000e+00 2.868e+00 2.274e-04
219 760.558 123.914 2.601e-02 7.046e-03 5.663e-22 1.000e+00 0.000e+00 2.867e+00 2.259e-04
220 761.802 123.750 2.584e-02 6.870e-03 5.311e-22 1.000e+00 0.000e+00 2.866e+00 2.243e-04
221 763.060 123.256 2.567e-02 6.729e-03 5.615e-22 1.000e+00 0.000e+00 2.864e+00 2.228e-04
222 764.310 122.509 2.550e-02 6.613e-03 6.061e-22 1.000e+00 0.000e+00 2.859e+00 2.213e-04
223 765.557 121.681 2.534e-02 6.524e-03 6.186e-22 1.000e+00 0.000e+00 2.851e+00 2.198e-04
224 766.815 121.062 2.517e-02 6.466e-03 5.746e-22 1.000e+00 0.000e+00 2.843e+00 2.184e-04
225 768.071 120.731 2.500e-02 6.448e-03 4.879e-22 1.000e+00 0.000e+00 2.834e+00 2.169e-04
226 769.326 120.550 2.484e-02 6.482e-03 3.904e-22 1.000e+00 0.000e+00 2.824e+00 2.155e-04
227 770.564 120.456 2.468e-02 6.585e-03 3.155e-22 1.000e+00 0.000e+00 2.813e+00 2.140e-04
228 771.823 120.093 2.452e-02 6.768e-03 2.764e-22 1.000e+00 0.000e+00 2.800e+00 2.126e-04
229 773.074 119.833 2.436e-02 7.035e-03 2.723e-22 1.000e+00 0.000e+00 2.786e+00 2.112e-04
230 774.338 119.604 2.420e-02 7.369e-03 2.949e-22 1.000e+00 0.000e+00 2.771e+00 2.098e-04
231 776.832 119.136 2.389e-02 8.069e-03 3.684e-22 1.000e+00 0.000e+00 2.741e+00 2.070e-04
232 779.336 118.623 2.358e-02 8.375e-03 4.135e-22 1.000e+00 0.000e+00 2.699e+00 2.043e-04
233 781.843 118.090 2.328e-02 7.962e-03 4.360e-22 1.000e+00 0.000e+00 2.650e+00 2.016e-04
234 784.350 117.590 2.298e-02 7.054e-03 4.865e-22 1.000e+00 0.000e+00 2.598e+00 1.990e-04
235 786.855 117.193 2.269e-02 6.138e-03 6.189e-22 1.000e+00 0.000e+00 2.541e+00 1.964e-04
236 789.367 116.427 2.240e-02 5.442e-03 8.418e-22 1.000e+00 0.000e+00 2.482e+00 1.939e-04
237 791.865 114.579 2.212e-02 4.901e-03 7.865e-22 1.000e+00 0.000e+00 2.423e+00 1.914e-04
238 794.382 113.306 2.184e-02 4.449e-03 4.479e-22 1.000e+00 0.000e+00 2.369e+00 1.889e-04
239 796.881 113.682 2.157e-02 4.130e-03 2.896e-22 1.000e+00 0.000e+00 2.321e+00 1.865e-04
240 799.394 113.560 2.130e-02 3.943e-03 3.150e-22 1.000e+00 0.000e+00 2.274e+00 1.841e-04
241 801.901 112.589 2.104e-02 3.847e-03 3.325e-22 1.000e+00 0.000e+00 2.236e+00 1.818e-04
242 804.409 111.657 2.078e-02 3.891e-03 2.312e-22 1.000e+00 0.000e+00 2.212e+00 1.794e-04
243 806.913 110.856 2.052e-02 4.171e-03 1.138e-22 1.000e+00 0.000e+00 2.196e+00 1.772e-04
244 809.428 110.319 2.027e-02 4.692e-03 9.675e-23 1.000e+00 0.000e+00 2.192e+00 1.750e-04
245 811.932 110.212 2.003e-02 5.313e-03 1.269e-22 1.000e+00 0.000e+00 2.203e+00 1.728e-04
246 814.440 109.950 1.979e-02 5.836e-03 1.403e-22 1.000e+00 0.000e+00 2.232e+00 1.706e-04
247 816.943 108.713 1.955e-02 6.024e-03 1.070e-22 1.000e+00 0.000e+00 2.276e+00 1.685e-04
248 819.456 107.005 1.932e-02 5.699e-03 6.941e-23 1.000e+00 0.000e+00 2.340e+00 1.665e-04
249 821.961 106.334 1.908e-02 4.916e-03 6.586e-23 1.000e+00 0.000e+00 2.440e+00 1.644e-04
250 824.462 106.615 1.885e-02 3.982e-03 8.298e-23 1.000e+00 0.000e+00 2.605e+00 1.623e-04
251 826.984 106.526 1.862e-02 3.185e-03 1.014e-22 1.000e+00 0.000e+00 2.847e+00 1.603e-04
252 829.489 105.761 1.840e-02 2.620e-03 1.147e-22 1.000e+00 0.000e+00 3.138e+00 1.583e-04
253 832.005 104.362 1.817e-02 2.259e-03 1.471e-22 1.000e+00 0.000e+00 3.434e+00 1.564e-04
254 834.507 103.505 1.795e-02 2.051e-03 1.970e-22 1.000e+00 0.000e+00 3.664e+00 1.544e-04
255 837.018 103.416 1.774e-02 1.974e-03 1.945e-22 1.000e+00 0.000e+00 3.810e+00 1.526e-04
256 839.516 103.189 1.754e-02 1.990e-03 1.761e-22 1.000e+00 0.000e+00 3.914e+00 1.508e-04
257 842.031 102.548 1.733e-02 2.081e-03 1.913e-22 1.000e+00 0.000e+00 3.996e+00 1.489e-04
258 844.538 101.894 1.713e-02 2.267e-03 1.738e-22 1.000e+00 0.000e+00 4.064e+00 1.471e-04
259 847.062 100.764 1.693e-02 2.602e-03 1.158e-22 1.000e+00 0.000e+00 4.128e+00 1.453e-04
260 849.572 98.537 1.673e-02 3.172e-03 7.480e-23 1.000e+00 0.000e+00 4.189e+00 1.436e-04
261 852.086 94.626 1.654e-02 3.781e-03 8.119e-23 1.000e+00 0.000e+00 4.243e+00 1.419e-04
262 854.586 92.389 1.634e-02 3.974e-03 1.025e-22 1.000e+00 0.000e+00 4.301e+00 1.402e-04
263 857.103 95.844 1.614e-02 3.669e-03 8.662e-23 1.000e+00 0.000e+00 4.366e+00 1.385e-04
264 859.594 98.543 1.595e-02 3.165e-03 6.504e-23 1.000e+00 0.000e+00 4.432e+00 1.368e-04
265 862.112 97.885 1.577e-02 2.572e-03 6.390e-23 1.000e+00 0.000e+00 4.501e+00 1.352e-04
266 864.610 94.383 1.560e-02 2.051e-03 5.998e-23 1.000e+00 0.000e+00 4.585e+00 1.336e-04
267 867.113 92.034 1.542e-02 1.649e-03 6.107e-23 1.000e+00 0.000e+00 4.670e+00 1.321e-04
268 869.613 94.195 1.524e-02 1.362e-03 6.062e-23 1.000e+00 0.000e+00 4.755e+00 1.305e-04
269 872.129 95.128 1.507e-02 1.178e-03 5.796e-23 1.000e+00 0.000e+00 4.860e+00 1.290e-04
270 874.636 94.383 1.490e-02 1.081e-03 6.097e-23 1.000e+00 0.000e+00 4.980e+00 1.275e-04
271 877.145 94.042 1.472e-02 1.057e-03 6.736e-23 1.000e+00 0.000e+00 5.110e+00 1.260e-04
272 879.637 93.568 1.456e-02 1.097e-03 7.289e-23 1.000e+00 0.000e+00 5.252e+00 1.245e-04
273 882.136 93.350 1.440e-02 1.184e-03 7.406e-23 1.000e+00 0.000e+00 5.399e+00 1.231e-04
274 884.639 93.096 1.424e-02 1.303e-03 7.027e-23 1.000e+00 0.000e+00 5.535e+00 1.217e-04
275 887.130 92.483 1.408e-02 1.422e-03 7.840e-23 1.000e+00 0.000e+00 5.669e+00 1.203e-04
276 889.598 92.305 1.393e-02 1.469e-03 1.132e-22 1.000e+00 0.000e+00 5.813e+00 1.190e-04
277 892.093 91.818 1.378e-02 1.484e-03 1.219e-22 1.000e+00 0.000e+00 5.950e+00 1.177e-04
278 894.601 91.425 1.364e-02 1.610e-03 9.420e-23 1.000e+00 0.000e+00 6.078e+00 1.164e-04
279 939.713 81.998 1.109e-02 5.011e-04 7.228e-24 1.000e+00 0.000e+00 2.247e+01 9.450e-05
280 1038.317 66.984 7.465e-03 3.476e-05 3.122e-27 1.000e+00 0.000e+00 2.308e+01 6.293e-05
281 1248.550 44.519 3.536e-03 8.538e-09 1.212e-26 1.000e+00 0.000e+00 1.125e+02 2.952e-05
282 1378.169 35.562 2.376e-03 1.117e-08 3.016e-26 1.000e+00 0.000e+00 5.588e+02 1.971e-05
283 1618.034 23.487 1.252e-03 3.024e-09 8.983e-27 1.000e+00 0.000e+00 7.136e+02 1.026e-05
284 2130.593 9.111 4.152e-04 1.149e-08 3.418e-26 1.000e+00 0.000e+00 2.521e+03 3.356e-06
285 2258.429 7.397 3.292e-04 9.446e-09 2.821e-26 1.000e+00 0.000e+00 2.208e+03 2.651e-06
Loading default parameters for OCI from /opt/ocssw/share/oci/msl12_defaults.par
Loading parameters for suite SFREFL from /opt/ocssw/share/common/msl12_defaults_SFREFL.par
Loading parameters for suite SFREFL from /opt/ocssw/share/oci/msl12_defaults_SFREFL.par
Loading command line parameters
Loading user parameters for OCI
Internal data compression requested at compression level: 4
Opening filter file /opt/ocssw/share/oci/msl12_filter.dat
Setting 5 x 3 straylight filter on CLDICE mask
Filter Kernel
1 1 1 1 1
1 1 1 1 1
1 1 1 1 1
Minimum fill set to 1 pixels
Opening OCI L1B file
OCI L1B Npix :1272 Nlines:1710
file->nbands = 286
Allocated 32313760 bytes in L1 record.
Allocated 13157568 bytes in L2 record.
Opening: data/PACE_OCI.20240605T092137.L2_SFREFL.V2.nc
The following products will be included in data/PACE_OCI.20240605T092137.L2_SFREFL.V2.nc.
0 rhos
1 l2_flags
Begin l2gen Version 9.7.0-V2024.1 Processing
Sensor is OCI
Sensor ID is 30
Sensor has 286 reflective bands
Sensor has 0 emissive bands
Number of along-track detectors per band is 1
Number of input pixels per scan is 1272
Processing pixels 1 to 1272 by 1
Processing scans 1 to 1710 by 1
Ocean processing enabled
Land processing enabled
Atmospheric correction disabled
Begin MSl12 processing at 2024217025421000
Allocated 32313760 bytes in L1 record.
Allocated 32313760 bytes in L1 record.
Allocated 32313760 bytes in L1 record.
Loading land mask information from /opt/ocssw/share/common/gebco_ocssw_v2020.nc
Loading DEM information from /opt/ocssw/share/common/gebco_ocssw_v2020.nc
Loading ice mask file from /opt/ocssw/share/common/ice_climatology.hdf
Loaded monthly NSIDC ice climatology HDF file.
Loading DEM info from /opt/ocssw/share/common/gebco_ocssw_v2020.nc
Loading climatology file /opt/ocssw/share/common/sst_climatology.hdf
Loading SSS reference from Climatology file: /opt/ocssw/share/common/sss_climatology_woa2009.hdf
Opening meteorological files.
met1 = /opt/ocssw/share/common/met_climatology_v2014.hdf
met2 =
met3 =
ozone1 = /opt/ocssw/share/common/ozone_climatology_v2014.hdf
ozone2 =
ozone3 =
no2 = /opt/ocssw/share/common/no2_climatology_v2013.hdf
Opening ozone file /opt/ocssw/share/common/ozone_climatology_v2014.hdf
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_315_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_316_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_318_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_320_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_322_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_325_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_327_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_329_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_331_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_334_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_337_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_339_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_341_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_344_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_346_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_348_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_351_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_353_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_356_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_358_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_361_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_363_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_366_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_368_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_371_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_373_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_375_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_378_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_380_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_383_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_385_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_388_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_390_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_393_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_395_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_398_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_400_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_403_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_405_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_408_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_410_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_413_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_415_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_418_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_420_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_422_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_425_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_427_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_430_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_432_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_435_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_437_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_440_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_442_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_445_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_447_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_450_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_452_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_455_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_457_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_460_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_462_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_465_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_467_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_470_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_472_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_475_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_477_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_480_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_482_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_485_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_487_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_490_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_492_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_495_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_497_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_500_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_502_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_505_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_507_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_510_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_512_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_515_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_517_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_520_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_522_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_525_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_527_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_530_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_532_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_535_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_537_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_540_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_542_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_545_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_547_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_550_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_553_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_555_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_558_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_560_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_563_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_565_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_568_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_570_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_573_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_575_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_578_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_580_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_583_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_586_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_588_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_591_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_593_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_596_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_598_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_601_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_603_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_605_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_608_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_610_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_613_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_615_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_618_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_620_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_623_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_625_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_627_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_630_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_632_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_635_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_637_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_640_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_641_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_642_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_643_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_645_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_646_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_647_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_648_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_650_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_651_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_652_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_653_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_655_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_656_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_657_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_658_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_660_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_661_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_662_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_663_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_665_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_666_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_667_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_668_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_670_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_671_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_672_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_673_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_675_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_676_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_677_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_678_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_679_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_681_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_682_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_683_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_684_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_686_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_687_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_688_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_689_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_691_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_692_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_693_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_694_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_696_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_697_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_698_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_699_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_701_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_702_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_703_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_704_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_706_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_707_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_708_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_709_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_711_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_712_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_713_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_714_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_717_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_719_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_722_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_724_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_727_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_729_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_732_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_734_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_737_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_739_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_741_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_742_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_743_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_744_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_746_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_747_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_748_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_749_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_751_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_752_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_753_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_754_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_756_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_757_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_758_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_759_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_761_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_762_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_763_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_764_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_766_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_767_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_768_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_769_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_771_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_772_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_773_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_774_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_777_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_779_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_782_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_784_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_787_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_789_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_792_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_794_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_797_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_799_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_802_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_804_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_807_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_809_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_812_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_814_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_817_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_819_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_822_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_824_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_827_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_829_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_832_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_835_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_837_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_840_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_842_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_845_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_847_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_850_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_852_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_855_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_857_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_860_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_862_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_865_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_867_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_870_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_872_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_875_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_877_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_880_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_882_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_885_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_887_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_890_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_892_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_895_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_940_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_1038_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_1249_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_1378_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_1618_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_2131_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_2258_iqu.nc
Using 869.6 nm channel for cloud flagging over water.
Using 412.6 nm channel for cloud flagging over land.
Processing scan # 0 (1 of 1710) after 10 seconds
Processing scan # 50 (51 of 1710) after 28 seconds
Processing scan # 100 (101 of 1710) after 46 seconds
Processing scan # 150 (151 of 1710) after 64 seconds
Processing scan # 200 (201 of 1710) after 81 seconds
Processing scan # 250 (251 of 1710) after 98 seconds
Processing scan # 300 (301 of 1710) after 114 seconds
Processing scan # 350 (351 of 1710) after 131 seconds
Processing scan # 400 (401 of 1710) after 147 seconds
Processing scan # 450 (451 of 1710) after 164 seconds
Processing scan # 500 (501 of 1710) after 181 seconds
Processing scan # 550 (551 of 1710) after 197 seconds
Processing scan # 600 (601 of 1710) after 213 seconds
Processing scan # 650 (651 of 1710) after 231 seconds
Processing scan # 700 (701 of 1710) after 247 seconds
Processing scan # 750 (751 of 1710) after 264 seconds
Processing scan # 800 (801 of 1710) after 282 seconds
Processing scan # 850 (851 of 1710) after 300 seconds
Processing scan # 900 (901 of 1710) after 319 seconds
Processing scan # 950 (951 of 1710) after 338 seconds
Processing scan # 1000 (1001 of 1710) after 357 seconds
Processing scan # 1050 (1051 of 1710) after 375 seconds
Processing scan # 1100 (1101 of 1710) after 393 seconds
Processing scan # 1150 (1151 of 1710) after 411 seconds
Processing scan # 1200 (1201 of 1710) after 429 seconds
Processing scan # 1250 (1251 of 1710) after 447 seconds
Processing scan # 1300 (1301 of 1710) after 464 seconds
Processing scan # 1350 (1351 of 1710) after 483 seconds
Processing scan # 1400 (1401 of 1710) after 502 seconds
Processing scan # 1450 (1451 of 1710) after 521 seconds
Processing scan # 1500 (1501 of 1710) after 540 seconds
Processing scan # 1550 (1551 of 1710) after 560 seconds
Processing scan # 1600 (1601 of 1710) after 580 seconds
Processing scan # 1650 (1651 of 1710) after 599 seconds
Processing scan # 1700 (1701 of 1710) after 617 seconds
Percentage of pixels flagged:
Flag # 1: ATMFAIL 0 0.0000
Flag # 2: LAND 1791549 82.3655
Flag # 3: PRODWARN 0 0.0000
Flag # 4: HIGLINT 14899 0.6850
Flag # 5: HILT 71193 3.2731
Flag # 6: HISATZEN 309756 14.2409
Flag # 7: COASTZ 121712 5.5956
Flag # 8: SPARE 0 0.0000
Flag # 9: STRAYLIGHT 286472 13.1704
Flag #10: CLDICE 750283 34.4939
Flag #11: COCCOLITH 0 0.0000
Flag #12: TURBIDW 0 0.0000
Flag #13: HISOLZEN 0 0.0000
Flag #14: SPARE 0 0.0000
Flag #15: LOWLW 0 0.0000
Flag #16: CHLFAIL 0 0.0000
Flag #17: NAVWARN 0 0.0000
Flag #18: ABSAER 0 0.0000
Flag #19: SPARE 0 0.0000
Flag #20: MAXAERITER 0 0.0000
Flag #21: MODGLINT 37871 1.7411
Flag #22: CHLWARN 0 0.0000
Flag #23: ATMWARN 0 0.0000
Flag #24: SPARE 0 0.0000
Flag #25: SEAICE 0 0.0000
Flag #26: NAVFAIL 0 0.0000
Flag #27: FILTER 0 0.0000
Flag #28: SPARE 0 0.0000
Flag #29: BOWTIEDEL 0 0.0000
Flag #30: HIPOL 0 0.0000
Flag #31: PRODFAIL 0 0.0000
Flag #32: SPARE 0 0.0000
End MSl12 processing at 2024217030442000
Processing Rate = 2.753623 scans/sec
Closing oci l1b file
Processing Completed
If successful, the l2gen program created a netCDF file at the ofile path. The contents should include the chlor_a product from the BGC suite of products. Once this process is done, you are ready to visualize your “custom” L2 data. Use the robust=True option to ignore outlier chl a values.
dataset = xr.open_dataset(par["ofile"], group="geophysical_data")
artist = dataset["rhos"].sel({"wavelength_3d": 25}).plot(cmap="viridis", robust=True)
Feel free to explore l2gen options to produce the Level-2 dataset you need! The documentation
for l2gen is kind of interactive, because so much depends on the data product being processed.
For example, try l2gen ifile=data/PACE_OCI.20240605T092137.L1B.V2.nc dump_options=true to get
a lot of information about the specifics of what the l2gen program generates.
The next step for this tutorial is to merge multiple Level-2 granules together.
4. Make MOANA Phytoplankton Community Products with l2gen#
One of the most exciting new PACE algorithms for the ocean color community is the MOANA algorithm that produces phytoplankton abundances for three different groups: Synechococcus, Prochlorococcus and picoeukaryotes.
tspan = ("2024-03-09", "2024-03-09")
location = (20, -30)
results = earthaccess.search_data(
short_name="PACE_OCI_L1B_SCI",
temporal=tspan,
point=location,
)
results[0]
Download the granules found in the search.
paths = earthaccess.download(results, local_path="data")
The Level-1B files contain top-of-atmosphere reflectances, typically denoted as \(\rho_t\).
On OCI, the reflectances are grouped into blue, red, and short-wave infrared (SWIR) wavelengths. Open
the dataset’s “observatin_data” group in the NetCDF file using xarray to plot a “rhot_red”
wavelength.
dataset = xr.open_dataset(paths[0], group="observation_data")
artist = dataset["rhot_red"].sel({"red_bands": 100}).plot()
ifile = paths[0]
par = {
"ifile": ifile,
"ofile": str(ifile).replace(".L1B.", ".L2_MOANA."),
"suite": "BGC",
"l2prod": "picoeuk_moana prococcus_moana syncoccus_moana rhos_465 rhos_555 rhos_645 poc chlor_a ",
"atmocor": 1,
}
write_par("l2gen-moana.par", par)
Now run l2bin using your chosen parameters. Be very patient.
%%bash
source $OCSSWROOT/OCSSW_bash.env
l2gen par=l2gen-moana.par
Loading default parameters from /opt/ocssw/share/common/msl12_defaults.par
Input file data/PACE_OCI.20240309T115927.L1B.V2.nc is PACE L1B file.
Loading characteristics for OCI
Opening sensor information file /opt/ocssw/share/oci/msl12_sensor_info.dat
Bnd Lam Fo Tau_r k_oz k_no2 t_co2 awhite aw bbw
0 314.550 112.026 4.873e-01 4.208e-01 3.281e-19 1.000e+00 0.000e+00 2.305e-01 6.356e-03
1 316.239 92.478 6.485e-01 5.806e-01 2.961e-19 1.000e+00 0.000e+00 1.633e-01 7.727e-03
2 318.262 85.195 7.410e-01 5.473e-01 2.844e-19 1.000e+00 0.000e+00 1.278e-01 8.187e-03
3 320.303 82.175 7.809e-01 4.609e-01 2.833e-19 1.000e+00 0.000e+00 1.105e-01 8.271e-03
4 322.433 80.733 7.906e-01 3.543e-01 2.898e-19 1.000e+00 0.000e+00 9.950e-02 8.190e-03
5 324.649 86.251 7.915e-01 2.567e-01 3.018e-19 1.000e+00 0.000e+00 9.079e-02 8.041e-03
6 326.828 95.932 7.891e-01 1.907e-01 3.132e-19 1.000e+00 0.000e+00 8.475e-02 7.871e-03
7 328.988 101.672 7.700e-01 1.386e-01 3.251e-19 1.000e+00 0.000e+00 8.211e-02 7.627e-03
8 331.305 101.708 7.404e-01 9.852e-02 3.417e-19 1.000e+00 0.000e+00 8.089e-02 7.342e-03
9 333.958 97.745 7.204e-01 6.830e-02 3.572e-19 1.000e+00 0.000e+00 7.656e-02 7.132e-03
10 336.815 93.497 7.112e-01 4.395e-02 3.736e-19 1.000e+00 0.000e+00 6.891e-02 6.975e-03
11 339.160 96.099 7.034e-01 3.070e-02 3.963e-19 1.000e+00 0.000e+00 6.288e-02 6.834e-03
12 341.321 98.332 6.898e-01 2.060e-02 4.076e-19 1.000e+00 0.000e+00 5.915e-02 6.663e-03
13 343.632 95.167 6.720e-01 1.439e-02 4.137e-19 1.000e+00 0.000e+00 5.624e-02 6.474e-03
14 346.017 92.958 6.529e-01 9.882e-03 4.395e-19 1.000e+00 0.000e+00 5.365e-02 6.285e-03
15 348.468 95.422 6.342e-01 6.032e-03 4.610e-19 1.000e+00 0.000e+00 5.106e-02 6.097e-03
16 350.912 99.423 6.163e-01 4.542e-03 4.617e-19 1.000e+00 0.000e+00 4.864e-02 5.913e-03
17 353.344 102.927 5.988e-01 3.473e-03 4.768e-19 1.000e+00 0.000e+00 4.647e-02 5.735e-03
18 355.782 99.180 5.823e-01 2.499e-03 5.019e-19 1.000e+00 0.000e+00 4.450e-02 5.562e-03
19 358.235 93.459 5.642e-01 2.283e-03 5.089e-19 1.000e+00 0.000e+00 4.256e-02 5.394e-03
20 360.695 97.255 5.472e-01 2.096e-03 5.117e-19 1.000e+00 0.000e+00 4.053e-02 5.233e-03
21 363.137 105.330 5.320e-01 1.734e-03 5.326e-19 1.000e+00 0.000e+00 3.856e-02 5.079e-03
22 365.610 113.914 5.178e-01 1.415e-03 5.533e-19 1.000e+00 0.000e+00 3.642e-02 4.932e-03
23 368.083 118.845 5.043e-01 1.282e-03 5.555e-19 1.000e+00 0.000e+00 3.417e-02 4.789e-03
24 370.534 115.549 4.908e-01 1.286e-03 5.590e-19 1.000e+00 0.000e+00 3.126e-02 4.651e-03
25 372.991 107.218 4.773e-01 1.342e-03 5.744e-19 1.000e+00 0.000e+00 2.719e-02 4.520e-03
26 375.482 108.880 4.630e-01 1.283e-03 5.834e-19 1.000e+00 0.000e+00 2.270e-02 4.392e-03
27 377.926 118.137 4.513e-01 1.115e-03 5.861e-19 1.000e+00 0.000e+00 1.843e-02 4.269e-03
28 380.419 111.060 4.406e-01 1.167e-03 5.945e-19 1.000e+00 0.000e+00 1.508e-02 4.151e-03
29 382.876 96.370 4.280e-01 1.379e-03 6.014e-19 1.000e+00 0.000e+00 1.316e-02 4.037e-03
30 385.359 96.688 4.156e-01 1.389e-03 6.039e-19 1.000e+00 0.000e+00 1.228e-02 3.928e-03
31 387.811 107.161 4.051e-01 1.257e-03 6.130e-19 1.000e+00 0.000e+00 1.166e-02 3.822e-03
32 390.297 112.501 3.955e-01 1.196e-03 6.227e-19 1.000e+00 0.000e+00 1.104e-02 3.718e-03
33 392.764 106.686 3.857e-01 1.256e-03 6.193e-19 1.000e+00 0.000e+00 1.065e-02 3.619e-03
34 395.238 106.058 3.747e-01 1.271e-03 6.157e-19 1.000e+00 0.000e+00 1.040e-02 3.523e-03
35 397.706 127.304 3.647e-01 1.082e-03 6.328e-19 1.000e+00 0.000e+00 9.891e-03 3.430e-03
36 400.178 157.144 3.564e-01 9.418e-04 6.342e-19 1.000e+00 0.000e+00 8.906e-03 3.341e-03
37 402.654 170.763 3.486e-01 9.296e-04 6.155e-19 1.000e+00 0.000e+00 7.976e-03 3.255e-03
38 405.127 168.889 3.400e-01 9.759e-04 6.015e-19 1.000e+00 0.000e+00 7.534e-03 3.171e-03
39 407.605 168.988 3.314e-01 1.065e-03 6.101e-19 1.000e+00 0.000e+00 7.203e-03 3.090e-03
40 410.074 173.399 3.233e-01 1.143e-03 6.226e-19 1.000e+00 0.000e+00 6.868e-03 3.011e-03
41 412.557 177.268 3.155e-01 1.158e-03 6.161e-19 1.000e+00 0.000e+00 6.656e-03 2.935e-03
42 415.025 177.759 3.079e-01 1.218e-03 5.856e-19 1.000e+00 0.000e+00 6.564e-03 2.861e-03
43 417.512 175.925 3.004e-01 1.325e-03 5.698e-19 1.000e+00 0.000e+00 6.661e-03 2.789e-03
44 419.988 175.156 2.932e-01 1.448e-03 5.927e-19 1.000e+00 0.000e+00 6.771e-03 2.720e-03
45 422.453 174.012 2.863e-01 1.774e-03 6.054e-19 1.000e+00 0.000e+00 6.878e-03 2.654e-03
46 424.940 170.766 2.796e-01 2.153e-03 5.793e-19 1.000e+00 0.000e+00 7.006e-03 2.590e-03
47 427.398 162.027 2.732e-01 2.242e-03 5.468e-19 1.000e+00 0.000e+00 6.986e-03 2.527e-03
48 429.885 154.088 2.664e-01 2.290e-03 5.331e-19 1.000e+00 0.000e+00 6.994e-03 2.464e-03
49 432.379 161.564 2.597e-01 2.492e-03 5.452e-19 1.000e+00 0.000e+00 7.152e-03 2.404e-03
50 434.869 174.172 2.539e-01 2.644e-03 5.549e-19 1.000e+00 0.000e+00 7.438e-03 2.347e-03
51 437.351 178.115 2.482e-01 3.023e-03 5.355e-19 1.000e+00 0.000e+00 8.014e-03 2.291e-03
52 439.828 182.229 2.424e-01 3.903e-03 4.968e-19 1.000e+00 0.000e+00 8.606e-03 2.238e-03
53 442.327 190.795 2.368e-01 4.600e-03 4.726e-19 1.000e+00 0.000e+00 9.139e-03 2.185e-03
54 444.811 195.628 2.315e-01 4.590e-03 4.945e-19 1.000e+00 0.000e+00 9.759e-03 2.133e-03
55 447.309 199.848 2.261e-01 4.547e-03 5.107e-19 1.000e+00 0.000e+00 1.052e-02 2.082e-03
56 449.795 205.764 2.211e-01 4.968e-03 4.798e-19 1.000e+00 0.000e+00 1.131e-02 2.034e-03
57 452.280 205.708 2.162e-01 5.368e-03 4.441e-19 1.000e+00 0.000e+00 1.179e-02 1.987e-03
58 454.769 204.450 2.114e-01 5.945e-03 4.252e-19 1.000e+00 0.000e+00 1.203e-02 1.942e-03
59 457.262 205.953 2.067e-01 7.420e-03 4.263e-19 1.000e+00 0.000e+00 1.212e-02 1.898e-03
60 459.757 207.117 2.022e-01 9.301e-03 4.351e-19 1.000e+00 0.000e+00 1.214e-02 1.855e-03
61 462.252 207.870 1.977e-01 1.011e-02 4.326e-19 1.000e+00 0.000e+00 1.219e-02 1.813e-03
62 464.737 205.886 1.935e-01 9.683e-03 4.061e-19 1.000e+00 0.000e+00 1.238e-02 1.772e-03
63 467.244 203.675 1.893e-01 9.647e-03 3.691e-19 1.000e+00 0.000e+00 1.242e-02 1.733e-03
64 469.729 203.713 1.852e-01 1.043e-02 3.441e-19 1.000e+00 0.000e+00 1.244e-02 1.694e-03
65 472.202 205.281 1.812e-01 1.115e-02 3.582e-19 1.000e+00 0.000e+00 1.276e-02 1.657e-03
66 474.700 207.058 1.774e-01 1.242e-02 3.801e-19 1.000e+00 0.000e+00 1.326e-02 1.621e-03
67 477.189 208.385 1.736e-01 1.528e-02 3.580e-19 1.000e+00 0.000e+00 1.393e-02 1.586e-03
68 479.689 209.553 1.700e-01 1.895e-02 3.338e-19 1.000e+00 0.000e+00 1.437e-02 1.552e-03
69 482.183 206.590 1.665e-01 2.122e-02 3.022e-19 1.000e+00 0.000e+00 1.472e-02 1.518e-03
70 484.689 195.340 1.631e-01 2.114e-02 2.725e-19 1.000e+00 0.000e+00 1.533e-02 1.485e-03
71 487.182 189.524 1.595e-01 2.040e-02 2.939e-19 1.000e+00 0.000e+00 1.612e-02 1.454e-03
72 489.674 195.470 1.562e-01 2.101e-02 3.044e-19 1.000e+00 0.000e+00 1.694e-02 1.423e-03
73 492.176 199.211 1.530e-01 2.224e-02 2.853e-19 1.000e+00 0.000e+00 1.789e-02 1.393e-03
74 494.686 200.274 1.499e-01 2.354e-02 2.822e-19 1.000e+00 0.000e+00 1.916e-02 1.364e-03
75 497.182 199.355 1.469e-01 2.613e-02 2.498e-19 1.000e+00 0.000e+00 2.072e-02 1.336e-03
76 499.688 194.720 1.440e-01 3.087e-02 2.022e-19 1.000e+00 0.000e+00 2.233e-02 1.309e-03
77 502.190 192.753 1.410e-01 3.665e-02 2.151e-19 1.000e+00 0.000e+00 2.438e-02 1.281e-03
78 504.695 195.557 1.382e-01 4.082e-02 2.384e-19 1.000e+00 0.000e+00 2.700e-02 1.255e-03
79 507.198 197.605 1.355e-01 4.179e-02 2.315e-19 1.000e+00 0.000e+00 3.003e-02 1.230e-03
80 509.720 196.520 1.328e-01 4.101e-02 2.296e-19 1.000e+00 0.000e+00 3.389e-02 1.206e-03
81 512.213 193.098 1.302e-01 4.110e-02 2.157e-19 1.000e+00 0.000e+00 3.784e-02 1.181e-03
82 514.729 185.769 1.277e-01 4.275e-02 1.772e-19 1.000e+00 0.000e+00 4.045e-02 1.157e-03
83 517.219 179.326 1.252e-01 4.504e-02 1.597e-19 1.000e+00 0.000e+00 4.179e-02 1.134e-03
84 519.747 182.521 1.227e-01 4.767e-02 1.620e-19 1.000e+00 0.000e+00 4.270e-02 1.111e-03
85 522.249 189.089 1.203e-01 5.134e-02 1.624e-19 1.000e+00 0.000e+00 4.337e-02 1.089e-03
86 524.771 188.823 1.180e-01 5.654e-02 1.755e-19 1.000e+00 0.000e+00 4.387e-02 1.067e-03
87 527.276 187.920 1.158e-01 6.292e-02 1.771e-19 1.000e+00 0.000e+00 4.454e-02 1.049e-03
88 529.798 191.821 1.137e-01 6.884e-02 1.592e-19 1.000e+00 0.000e+00 4.553e-02 1.031e-03
89 532.314 193.286 1.114e-01 7.264e-02 1.423e-19 1.000e+00 0.000e+00 4.646e-02 1.008e-03
90 534.859 191.915 1.092e-01 7.422e-02 1.168e-19 1.000e+00 0.000e+00 4.742e-02 9.861e-04
91 537.346 190.229 1.072e-01 7.488e-02 1.024e-19 1.000e+00 0.000e+00 4.852e-02 9.673e-04
92 539.878 187.754 1.052e-01 7.665e-02 1.097e-19 1.000e+00 0.000e+00 4.966e-02 9.489e-04
93 542.395 188.298 1.032e-01 7.989e-02 1.240e-19 1.000e+00 0.000e+00 5.116e-02 9.303e-04
94 544.904 189.892 1.013e-01 8.325e-02 1.296e-19 1.000e+00 0.000e+00 5.328e-02 9.125e-04
95 547.441 189.629 9.940e-02 8.591e-02 1.237e-19 1.000e+00 0.000e+00 5.584e-02 8.953e-04
96 549.994 189.342 9.756e-02 8.809e-02 1.129e-19 1.000e+00 0.000e+00 5.851e-02 8.784e-04
97 552.511 189.725 9.575e-02 9.044e-02 9.907e-20 1.000e+00 0.000e+00 6.062e-02 8.617e-04
98 555.044 188.862 9.400e-02 9.359e-02 8.268e-20 1.000e+00 0.000e+00 6.191e-02 8.454e-04
99 557.576 185.867 9.230e-02 9.815e-02 6.751e-20 1.000e+00 0.000e+00 6.298e-02 8.297e-04
100 560.104 184.284 9.062e-02 1.041e-01 6.704e-20 1.000e+00 0.000e+00 6.443e-02 8.146e-04
101 562.642 184.903 8.899e-02 1.101e-01 8.271e-20 1.000e+00 0.000e+00 6.592e-02 7.997e-04
102 565.190 184.929 8.737e-02 1.152e-01 8.817e-20 1.000e+00 0.000e+00 6.740e-02 7.846e-04
103 567.710 184.394 8.579e-02 1.194e-01 8.097e-20 1.000e+00 0.000e+00 6.954e-02 7.698e-04
104 570.259 184.337 8.424e-02 1.231e-01 8.258e-20 1.000e+00 0.000e+00 7.243e-02 7.555e-04
105 572.796 185.701 8.273e-02 1.258e-01 7.445e-20 1.000e+00 0.000e+00 7.601e-02 7.418e-04
106 575.343 185.860 8.130e-02 1.262e-01 5.358e-20 1.000e+00 0.000e+00 8.077e-02 7.290e-04
107 577.902 184.295 7.992e-02 1.245e-01 4.606e-20 1.000e+00 0.000e+00 8.675e-02 7.169e-04
108 580.450 184.370 7.850e-02 1.217e-01 4.637e-20 1.000e+00 0.000e+00 9.411e-02 7.037e-04
109 582.996 184.516 7.711e-02 1.191e-01 4.563e-20 1.000e+00 0.000e+00 1.036e-01 6.904e-04
110 585.553 182.261 7.581e-02 1.175e-01 4.880e-20 1.000e+00 0.000e+00 1.147e-01 6.784e-04
111 588.086 178.467 7.455e-02 1.172e-01 5.159e-20 1.000e+00 0.000e+00 1.271e-01 6.670e-04
112 590.548 177.770 7.336e-02 1.186e-01 5.765e-20 1.000e+00 0.000e+00 1.411e-01 6.570e-04
113 593.084 179.810 7.228e-02 1.222e-01 5.611e-20 1.000e+00 0.000e+00 1.572e-01 6.487e-04
114 595.679 179.992 7.117e-02 1.275e-01 4.249e-20 1.000e+00 0.000e+00 1.772e-01 6.390e-04
115 598.262 178.361 6.999e-02 1.328e-01 3.893e-20 1.000e+00 0.000e+00 2.030e-01 6.276e-04
116 600.545 176.370 6.757e-02 1.365e-01 3.874e-20 1.000e+00 0.000e+00 2.467e-01 5.992e-04
117 602.920 176.174 6.651e-02 1.374e-01 3.034e-20 1.000e+00 0.000e+00 2.611e-01 5.899e-04
118 605.461 176.473 6.543e-02 1.353e-01 2.076e-20 1.000e+00 0.000e+00 2.702e-01 5.801e-04
119 607.986 174.705 6.437e-02 1.310e-01 1.990e-20 1.000e+00 0.000e+00 2.728e-01 5.706e-04
120 610.360 172.381 6.335e-02 1.256e-01 2.700e-20 1.000e+00 0.000e+00 2.733e-01 5.614e-04
121 612.730 170.269 6.236e-02 1.202e-01 3.487e-20 1.000e+00 0.000e+00 2.746e-01 5.524e-04
122 615.145 167.881 6.137e-02 1.152e-01 3.541e-20 1.000e+00 0.000e+00 2.763e-01 5.434e-04
123 617.605 167.707 6.038e-02 1.107e-01 3.180e-20 1.000e+00 0.000e+00 2.790e-01 5.346e-04
124 620.061 168.940 5.942e-02 1.070e-01 2.798e-20 1.000e+00 0.000e+00 2.832e-01 5.259e-04
125 622.530 167.529 5.848e-02 1.038e-01 2.439e-20 1.000e+00 0.000e+00 2.876e-01 5.173e-04
126 624.988 165.875 5.754e-02 1.007e-01 2.069e-20 1.000e+00 0.000e+00 2.918e-01 5.089e-04
127 627.434 166.459 5.663e-02 9.740e-02 1.634e-20 1.000e+00 0.000e+00 2.963e-01 5.006e-04
128 629.898 165.893 5.575e-02 9.400e-02 1.319e-20 1.000e+00 0.000e+00 3.008e-01 4.925e-04
129 632.376 164.212 5.486e-02 9.046e-02 1.304e-20 1.000e+00 0.000e+00 3.055e-01 4.846e-04
130 634.830 163.727 5.400e-02 8.676e-02 1.402e-20 1.000e+00 0.000e+00 3.097e-01 4.768e-04
131 637.305 163.541 5.316e-02 8.287e-02 1.464e-20 1.000e+00 0.000e+00 3.137e-01 4.692e-04
132 639.791 162.086 5.234e-02 7.890e-02 1.583e-20 1.000e+00 0.000e+00 3.191e-01 4.618e-04
133 641.029 161.259 5.193e-02 7.696e-02 1.681e-20 1.000e+00 0.000e+00 3.224e-01 4.581e-04
134 642.255 160.668 5.152e-02 7.512e-02 1.767e-20 1.000e+00 0.000e+00 3.258e-01 4.545e-04
135 643.479 160.372 5.112e-02 7.339e-02 1.849e-20 1.000e+00 0.000e+00 3.293e-01 4.509e-04
136 644.716 160.231 5.073e-02 7.176e-02 1.919e-20 1.000e+00 0.000e+00 3.327e-01 4.473e-04
137 645.966 159.889 5.034e-02 7.021e-02 1.946e-20 1.000e+00 0.000e+00 3.360e-01 4.437e-04
138 647.188 159.260 4.995e-02 6.871e-02 1.898e-20 1.000e+00 0.000e+00 3.396e-01 4.402e-04
139 648.435 158.731 4.956e-02 6.727e-02 1.756e-20 1.000e+00 0.000e+00 3.436e-01 4.367e-04
140 649.667 158.341 4.918e-02 6.588e-02 1.544e-20 1.000e+00 0.000e+00 3.486e-01 4.333e-04
141 650.913 158.275 4.880e-02 6.453e-02 1.328e-20 1.000e+00 0.000e+00 3.546e-01 4.299e-04
142 652.153 157.789 4.843e-02 6.321e-02 1.167e-20 1.000e+00 0.000e+00 3.616e-01 4.265e-04
143 653.388 155.177 4.807e-02 6.193e-02 1.068e-20 1.000e+00 0.000e+00 3.693e-01 4.231e-04
144 654.622 151.698 4.771e-02 6.060e-02 9.722e-21 1.000e+00 0.000e+00 3.774e-01 4.198e-04
145 655.869 148.463 4.733e-02 5.922e-02 8.805e-21 1.000e+00 0.000e+00 3.858e-01 4.166e-04
146 657.101 147.695 4.695e-02 5.782e-02 7.987e-21 1.000e+00 0.000e+00 3.947e-01 4.133e-04
147 658.340 149.658 4.658e-02 5.648e-02 7.170e-21 1.000e+00 0.000e+00 4.036e-01 4.101e-04
148 659.600 152.493 4.623e-02 5.523e-02 6.329e-21 1.000e+00 0.000e+00 4.119e-01 4.070e-04
149 660.833 154.868 4.590e-02 5.404e-02 5.541e-21 1.000e+00 0.000e+00 4.192e-01 4.038e-04
150 662.067 155.409 4.555e-02 5.283e-02 4.976e-21 1.000e+00 0.000e+00 4.251e-01 4.007e-04
151 663.300 155.290 4.521e-02 5.158e-02 4.897e-21 1.000e+00 0.000e+00 4.297e-01 3.976e-04
152 664.564 155.005 4.487e-02 5.029e-02 5.478e-21 1.000e+00 0.000e+00 4.334e-01 3.945e-04
153 665.795 154.711 4.453e-02 4.895e-02 6.640e-21 1.000e+00 0.000e+00 4.363e-01 3.915e-04
154 667.023 154.401 4.420e-02 4.759e-02 7.980e-21 1.000e+00 0.000e+00 4.389e-01 3.885e-04
155 668.263 154.028 4.387e-02 4.622e-02 9.022e-21 1.000e+00 0.000e+00 4.415e-01 3.855e-04
156 669.518 153.439 4.354e-02 4.488e-02 9.638e-21 1.000e+00 0.000e+00 4.442e-01 3.825e-04
157 670.755 152.834 4.321e-02 4.356e-02 9.943e-21 1.000e+00 0.000e+00 4.471e-01 3.796e-04
158 671.990 152.273 4.289e-02 4.229e-02 1.010e-20 1.000e+00 0.000e+00 4.497e-01 3.767e-04
159 673.245 151.870 4.257e-02 4.107e-02 1.027e-20 1.000e+00 0.000e+00 4.523e-01 3.738e-04
160 674.503 151.619 4.225e-02 3.991e-02 1.027e-20 1.000e+00 0.000e+00 4.551e-01 3.710e-04
161 675.731 151.421 4.194e-02 3.879e-02 9.926e-21 1.000e+00 0.000e+00 4.583e-01 3.681e-04
162 676.963 151.155 4.163e-02 3.774e-02 9.232e-21 1.000e+00 0.000e+00 4.620e-01 3.654e-04
163 678.208 150.791 4.132e-02 3.675e-02 8.304e-21 1.000e+00 0.000e+00 4.661e-01 3.626e-04
164 679.448 150.430 4.102e-02 3.584e-02 7.379e-21 1.000e+00 0.000e+00 4.703e-01 3.599e-04
165 680.680 149.905 4.072e-02 3.503e-02 6.552e-21 1.000e+00 0.000e+00 4.747e-01 3.572e-04
166 681.919 149.251 4.042e-02 3.430e-02 5.895e-21 1.000e+00 0.000e+00 4.794e-01 3.545e-04
167 683.171 148.503 4.012e-02 3.361e-02 5.490e-21 1.000e+00 0.000e+00 4.845e-01 3.518e-04
168 684.417 147.884 3.983e-02 3.292e-02 5.184e-21 1.000e+00 0.000e+00 4.901e-01 3.492e-04
169 685.657 147.515 3.953e-02 3.216e-02 4.905e-21 1.000e+00 0.000e+00 4.962e-01 3.466e-04
170 686.894 147.425 3.924e-02 3.131e-02 4.525e-21 1.000e+00 0.000e+00 5.029e-01 3.440e-04
171 688.143 147.400 3.896e-02 3.039e-02 4.024e-21 1.000e+00 0.000e+00 5.103e-01 3.414e-04
172 689.394 147.153 3.867e-02 2.943e-02 3.539e-21 1.000e+00 0.000e+00 5.186e-01 3.389e-04
173 690.647 146.715 3.839e-02 2.845e-02 3.144e-21 1.000e+00 0.000e+00 5.276e-01 3.363e-04
174 691.888 146.143 3.811e-02 2.750e-02 2.906e-21 1.000e+00 0.000e+00 5.375e-01 3.338e-04
175 693.130 145.684 3.784e-02 2.657e-02 2.763e-21 1.000e+00 0.000e+00 5.484e-01 3.314e-04
176 694.382 145.196 3.756e-02 2.567e-02 2.656e-21 1.000e+00 0.000e+00 5.605e-01 3.289e-04
177 695.644 144.701 3.729e-02 2.479e-02 2.542e-21 1.000e+00 0.000e+00 5.740e-01 3.265e-04
178 696.891 144.099 3.702e-02 2.394e-02 2.429e-21 1.000e+00 0.000e+00 5.888e-01 3.241e-04
179 698.118 143.280 3.676e-02 2.315e-02 2.343e-21 1.000e+00 0.000e+00 6.047e-01 3.217e-04
180 699.376 142.397 3.649e-02 2.241e-02 2.369e-21 1.000e+00 0.000e+00 6.215e-01 3.193e-04
181 700.612 141.532 3.623e-02 2.174e-02 2.581e-21 1.000e+00 0.000e+00 6.393e-01 3.170e-04
182 701.858 141.039 3.597e-02 2.114e-02 3.030e-21 1.000e+00 0.000e+00 6.580e-01 3.147e-04
183 703.097 140.869 3.571e-02 2.061e-02 3.660e-21 1.000e+00 0.000e+00 6.780e-01 3.124e-04
184 704.354 140.969 3.546e-02 2.014e-02 4.374e-21 1.000e+00 0.000e+00 6.995e-01 3.101e-04
185 705.593 140.996 3.521e-02 1.972e-02 4.920e-21 1.000e+00 0.000e+00 7.231e-01 3.078e-04
186 706.833 140.760 3.496e-02 1.934e-02 5.156e-21 1.000e+00 0.000e+00 7.499e-01 3.056e-04
187 708.089 140.307 3.471e-02 1.900e-02 5.111e-21 1.000e+00 0.000e+00 7.805e-01 3.034e-04
188 709.337 139.705 3.446e-02 1.868e-02 4.920e-21 1.000e+00 0.000e+00 8.152e-01 3.012e-04
189 710.581 139.142 3.422e-02 1.842e-02 4.776e-21 1.000e+00 0.000e+00 8.540e-01 2.990e-04
190 711.826 138.503 3.398e-02 1.820e-02 4.601e-21 1.000e+00 0.000e+00 8.959e-01 2.968e-04
191 713.068 137.918 3.374e-02 1.804e-02 4.294e-21 1.000e+00 0.000e+00 9.408e-01 2.947e-04
192 714.316 137.373 3.350e-02 1.788e-02 3.751e-21 1.000e+00 0.000e+00 9.886e-01 2.926e-04
193 716.817 136.139 3.303e-02 1.733e-02 2.447e-21 1.000e+00 0.000e+00 1.093e+00 2.884e-04
194 719.298 134.747 3.257e-02 1.625e-02 1.823e-21 1.000e+00 0.000e+00 1.206e+00 2.843e-04
195 721.800 134.248 3.212e-02 1.496e-02 1.858e-21 1.000e+00 0.000e+00 1.327e+00 2.802e-04
196 724.303 134.303 3.167e-02 1.391e-02 1.678e-21 1.000e+00 0.000e+00 1.461e+00 2.763e-04
197 726.796 133.424 3.124e-02 1.304e-02 1.167e-21 1.000e+00 0.000e+00 1.642e+00 2.724e-04
198 729.299 132.173 3.081e-02 1.220e-02 8.984e-22 1.000e+00 0.000e+00 1.891e+00 2.686e-04
199 731.790 131.770 3.038e-02 1.153e-02 9.392e-22 1.000e+00 0.000e+00 2.176e+00 2.648e-04
200 734.281 131.420 2.997e-02 1.114e-02 1.157e-21 1.000e+00 0.000e+00 2.438e+00 2.611e-04
201 736.791 130.150 2.956e-02 1.096e-02 1.301e-21 1.000e+00 0.000e+00 2.621e+00 2.575e-04
202 739.287 128.259 2.916e-02 1.090e-02 1.303e-21 1.000e+00 0.000e+00 2.732e+00 2.539e-04
203 740.535 127.659 2.897e-02 1.094e-02 1.339e-21 1.000e+00 0.000e+00 2.770e+00 2.521e-04
204 741.785 127.339 2.877e-02 1.103e-02 1.496e-21 1.000e+00 0.000e+00 2.799e+00 2.504e-04
205 743.046 127.458 2.857e-02 1.119e-02 1.813e-21 1.000e+00 0.000e+00 2.819e+00 2.486e-04
206 744.286 127.660 2.838e-02 1.137e-02 2.155e-21 1.000e+00 0.000e+00 2.832e+00 2.469e-04
207 745.534 127.796 2.819e-02 1.153e-02 2.323e-21 1.000e+00 0.000e+00 2.840e+00 2.452e-04
208 746.789 127.697 2.800e-02 1.159e-02 2.205e-21 1.000e+00 0.000e+00 2.846e+00 2.435e-04
209 748.041 127.310 2.781e-02 1.150e-02 1.876e-21 1.000e+00 0.000e+00 2.850e+00 2.419e-04
210 749.279 126.838 2.762e-02 1.122e-02 1.554e-21 1.000e+00 0.000e+00 2.854e+00 2.402e-04
211 750.540 126.351 2.744e-02 1.074e-02 1.390e-21 1.000e+00 0.000e+00 2.857e+00 2.386e-04
212 751.792 126.028 2.725e-02 1.013e-02 1.397e-21 1.000e+00 0.000e+00 2.862e+00 2.369e-04
213 753.042 125.814 2.707e-02 9.483e-03 1.443e-21 1.000e+00 0.000e+00 2.867e+00 2.353e-04
214 754.294 125.571 2.689e-02 8.866e-03 1.420e-21 1.000e+00 0.000e+00 2.870e+00 2.337e-04
215 755.542 125.152 2.671e-02 8.328e-03 1.307e-21 1.000e+00 0.000e+00 2.872e+00 2.321e-04
216 756.802 124.719 2.654e-02 7.888e-03 1.121e-21 1.000e+00 0.000e+00 2.871e+00 2.305e-04
217 758.051 124.328 2.636e-02 7.536e-03 8.963e-22 1.000e+00 0.000e+00 2.869e+00 2.290e-04
218 759.299 124.043 2.618e-02 7.263e-03 6.951e-22 1.000e+00 0.000e+00 2.868e+00 2.274e-04
219 760.558 123.914 2.601e-02 7.046e-03 5.663e-22 1.000e+00 0.000e+00 2.867e+00 2.259e-04
220 761.802 123.750 2.584e-02 6.870e-03 5.311e-22 1.000e+00 0.000e+00 2.866e+00 2.243e-04
221 763.060 123.256 2.567e-02 6.729e-03 5.615e-22 1.000e+00 0.000e+00 2.864e+00 2.228e-04
222 764.310 122.509 2.550e-02 6.613e-03 6.061e-22 1.000e+00 0.000e+00 2.859e+00 2.213e-04
223 765.557 121.681 2.534e-02 6.524e-03 6.186e-22 1.000e+00 0.000e+00 2.851e+00 2.198e-04
224 766.815 121.062 2.517e-02 6.466e-03 5.746e-22 1.000e+00 0.000e+00 2.843e+00 2.184e-04
225 768.071 120.731 2.500e-02 6.448e-03 4.879e-22 1.000e+00 0.000e+00 2.834e+00 2.169e-04
226 769.326 120.550 2.484e-02 6.482e-03 3.904e-22 1.000e+00 0.000e+00 2.824e+00 2.155e-04
227 770.564 120.456 2.468e-02 6.585e-03 3.155e-22 1.000e+00 0.000e+00 2.813e+00 2.140e-04
228 771.823 120.093 2.452e-02 6.768e-03 2.764e-22 1.000e+00 0.000e+00 2.800e+00 2.126e-04
229 773.074 119.833 2.436e-02 7.035e-03 2.723e-22 1.000e+00 0.000e+00 2.786e+00 2.112e-04
230 774.338 119.604 2.420e-02 7.369e-03 2.949e-22 1.000e+00 0.000e+00 2.771e+00 2.098e-04
231 776.832 119.136 2.389e-02 8.069e-03 3.684e-22 1.000e+00 0.000e+00 2.741e+00 2.070e-04
232 779.336 118.623 2.358e-02 8.375e-03 4.135e-22 1.000e+00 0.000e+00 2.699e+00 2.043e-04
233 781.843 118.090 2.328e-02 7.962e-03 4.360e-22 1.000e+00 0.000e+00 2.650e+00 2.016e-04
234 784.350 117.590 2.298e-02 7.054e-03 4.865e-22 1.000e+00 0.000e+00 2.598e+00 1.990e-04
235 786.855 117.193 2.269e-02 6.138e-03 6.189e-22 1.000e+00 0.000e+00 2.541e+00 1.964e-04
236 789.367 116.427 2.240e-02 5.442e-03 8.418e-22 1.000e+00 0.000e+00 2.482e+00 1.939e-04
237 791.865 114.579 2.212e-02 4.901e-03 7.865e-22 1.000e+00 0.000e+00 2.423e+00 1.914e-04
238 794.382 113.306 2.184e-02 4.449e-03 4.479e-22 1.000e+00 0.000e+00 2.369e+00 1.889e-04
239 796.881 113.682 2.157e-02 4.130e-03 2.896e-22 1.000e+00 0.000e+00 2.321e+00 1.865e-04
240 799.394 113.560 2.130e-02 3.943e-03 3.150e-22 1.000e+00 0.000e+00 2.274e+00 1.841e-04
241 801.901 112.589 2.104e-02 3.847e-03 3.325e-22 1.000e+00 0.000e+00 2.236e+00 1.818e-04
242 804.409 111.657 2.078e-02 3.891e-03 2.312e-22 1.000e+00 0.000e+00 2.212e+00 1.794e-04
243 806.913 110.856 2.052e-02 4.171e-03 1.138e-22 1.000e+00 0.000e+00 2.196e+00 1.772e-04
244 809.428 110.319 2.027e-02 4.692e-03 9.675e-23 1.000e+00 0.000e+00 2.192e+00 1.750e-04
245 811.932 110.212 2.003e-02 5.313e-03 1.269e-22 1.000e+00 0.000e+00 2.203e+00 1.728e-04
246 814.440 109.950 1.979e-02 5.836e-03 1.403e-22 1.000e+00 0.000e+00 2.232e+00 1.706e-04
247 816.943 108.713 1.955e-02 6.024e-03 1.070e-22 1.000e+00 0.000e+00 2.276e+00 1.685e-04
248 819.456 107.005 1.932e-02 5.699e-03 6.941e-23 1.000e+00 0.000e+00 2.340e+00 1.665e-04
249 821.961 106.334 1.908e-02 4.916e-03 6.586e-23 1.000e+00 0.000e+00 2.440e+00 1.644e-04
250 824.462 106.615 1.885e-02 3.982e-03 8.298e-23 1.000e+00 0.000e+00 2.605e+00 1.623e-04
251 826.984 106.526 1.862e-02 3.185e-03 1.014e-22 1.000e+00 0.000e+00 2.847e+00 1.603e-04
252 829.489 105.761 1.840e-02 2.620e-03 1.147e-22 1.000e+00 0.000e+00 3.138e+00 1.583e-04
253 832.005 104.362 1.817e-02 2.259e-03 1.471e-22 1.000e+00 0.000e+00 3.434e+00 1.564e-04
254 834.507 103.505 1.795e-02 2.051e-03 1.970e-22 1.000e+00 0.000e+00 3.664e+00 1.544e-04
255 837.018 103.416 1.774e-02 1.974e-03 1.945e-22 1.000e+00 0.000e+00 3.810e+00 1.526e-04
256 839.516 103.189 1.754e-02 1.990e-03 1.761e-22 1.000e+00 0.000e+00 3.914e+00 1.508e-04
257 842.031 102.548 1.733e-02 2.081e-03 1.913e-22 1.000e+00 0.000e+00 3.996e+00 1.489e-04
258 844.538 101.894 1.713e-02 2.267e-03 1.738e-22 1.000e+00 0.000e+00 4.064e+00 1.471e-04
259 847.062 100.764 1.693e-02 2.602e-03 1.158e-22 1.000e+00 0.000e+00 4.128e+00 1.453e-04
260 849.572 98.537 1.673e-02 3.172e-03 7.480e-23 1.000e+00 0.000e+00 4.189e+00 1.436e-04
261 852.086 94.626 1.654e-02 3.781e-03 8.119e-23 1.000e+00 0.000e+00 4.243e+00 1.419e-04
262 854.586 92.389 1.634e-02 3.974e-03 1.025e-22 1.000e+00 0.000e+00 4.301e+00 1.402e-04
263 857.103 95.844 1.614e-02 3.669e-03 8.662e-23 1.000e+00 0.000e+00 4.366e+00 1.385e-04
264 859.594 98.543 1.595e-02 3.165e-03 6.504e-23 1.000e+00 0.000e+00 4.432e+00 1.368e-04
265 862.112 97.885 1.577e-02 2.572e-03 6.390e-23 1.000e+00 0.000e+00 4.501e+00 1.352e-04
266 864.610 94.383 1.560e-02 2.051e-03 5.998e-23 1.000e+00 0.000e+00 4.585e+00 1.336e-04
267 867.113 92.034 1.542e-02 1.649e-03 6.107e-23 1.000e+00 0.000e+00 4.670e+00 1.321e-04
268 869.613 94.195 1.524e-02 1.362e-03 6.062e-23 1.000e+00 0.000e+00 4.755e+00 1.305e-04
269 872.129 95.128 1.507e-02 1.178e-03 5.796e-23 1.000e+00 0.000e+00 4.860e+00 1.290e-04
270 874.636 94.383 1.490e-02 1.081e-03 6.097e-23 1.000e+00 0.000e+00 4.980e+00 1.275e-04
271 877.145 94.042 1.472e-02 1.057e-03 6.736e-23 1.000e+00 0.000e+00 5.110e+00 1.260e-04
272 879.637 93.568 1.456e-02 1.097e-03 7.289e-23 1.000e+00 0.000e+00 5.252e+00 1.245e-04
273 882.136 93.350 1.440e-02 1.184e-03 7.406e-23 1.000e+00 0.000e+00 5.399e+00 1.231e-04
274 884.639 93.096 1.424e-02 1.303e-03 7.027e-23 1.000e+00 0.000e+00 5.535e+00 1.217e-04
275 887.130 92.483 1.408e-02 1.422e-03 7.840e-23 1.000e+00 0.000e+00 5.669e+00 1.203e-04
276 889.598 92.305 1.393e-02 1.469e-03 1.132e-22 1.000e+00 0.000e+00 5.813e+00 1.190e-04
277 892.093 91.818 1.378e-02 1.484e-03 1.219e-22 1.000e+00 0.000e+00 5.950e+00 1.177e-04
278 894.601 91.425 1.364e-02 1.610e-03 9.420e-23 1.000e+00 0.000e+00 6.078e+00 1.164e-04
279 939.713 81.998 1.109e-02 5.011e-04 7.228e-24 1.000e+00 0.000e+00 2.247e+01 9.450e-05
280 1038.317 66.984 7.465e-03 3.476e-05 3.122e-27 1.000e+00 0.000e+00 2.308e+01 6.293e-05
281 1248.550 44.519 3.536e-03 8.538e-09 1.212e-26 1.000e+00 0.000e+00 1.125e+02 2.952e-05
282 1378.169 35.562 2.376e-03 1.117e-08 3.016e-26 1.000e+00 0.000e+00 5.588e+02 1.971e-05
283 1618.034 23.487 1.252e-03 3.024e-09 8.983e-27 1.000e+00 0.000e+00 7.136e+02 1.026e-05
284 2130.593 9.111 4.152e-04 1.149e-08 3.418e-26 1.000e+00 0.000e+00 2.521e+03 3.356e-06
285 2258.429 7.397 3.292e-04 9.446e-09 2.821e-26 1.000e+00 0.000e+00 2.208e+03 2.651e-06
Loading default parameters for OCI from /opt/ocssw/share/oci/msl12_defaults.par
Loading parameters for suite BGC from /opt/ocssw/share/oci/msl12_defaults_BGC.par
Loading command line parameters
Loading user parameters for OCI
Internal data compression requested at compression level: 4
Opening filter file /opt/ocssw/share/oci/msl12_filter.dat
Setting 5 x 3 straylight filter on CLDICE mask
Filter Kernel
1 1 1 1 1
1 1 1 1 1
1 1 1 1 1
Minimum fill set to 1 pixels
Opening OCI L1B file
OCI L1B Npix :1272 Nlines:1710
file->nbands = 286
Allocated 32313760 bytes in L1 record.
Allocated 14633960 bytes in error record.
Allocated 16067904 bytes in L2 record.
Opening: data/PACE_OCI.20240309T115927.L2_MOANA.V2.nc
The following products will be included in data/PACE_OCI.20240309T115927.L2_MOANA.V2.nc.
0 picoeuk_moana
1 prococcus_moana
2 syncoccus_moana
3 rhos_465
4 rhos_555
5 rhos_645
6 poc
7 chlor_a
8 l2_flags
Begin l2gen Version 9.7.0-V2024.1 Processing
Sensor is OCI
Sensor ID is 30
Sensor has 286 reflective bands
Sensor has 0 emissive bands
Number of along-track detectors per band is 1
Number of input pixels per scan is 1272
Processing pixels 1 to 1272 by 1
Processing scans 1 to 1710 by 1
Ocean processing enabled
Land processing enabled
Atmospheric correction enabled
Begin MSl12 processing at 2024217030520000
Allocated 32313760 bytes in L1 record.
Allocated 32313760 bytes in L1 record.
Allocated 32313760 bytes in L1 record.
Allocated 14633960 bytes in error record.
Allocated 14633960 bytes in error record.
Allocated 14633960 bytes in error record.
Loading land mask information from /opt/ocssw/share/common/gebco_ocssw_v2020.nc
Loading DEM information from /opt/ocssw/share/common/gebco_ocssw_v2020.nc
Loading ice mask file from /opt/ocssw/share/common/ice_climatology.hdf
Loaded monthly NSIDC ice climatology HDF file.
Loading DEM info from /opt/ocssw/share/common/gebco_ocssw_v2020.nc
Loading climatology file /opt/ocssw/share/common/sst_climatology.hdf
Loading SSS reference from Climatology file: /opt/ocssw/share/common/sss_climatology_woa2009.hdf
Opening meteorological files.
met1 = /opt/ocssw/share/common/met_climatology_v2014.hdf
met2 =
met3 =
ozone1 = /opt/ocssw/share/common/ozone_climatology_v2014.hdf
ozone2 =
ozone3 =
no2 = /opt/ocssw/share/common/no2_climatology_v2013.hdf
Opening ozone file /opt/ocssw/share/common/ozone_climatology_v2014.hdf
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_315_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_316_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_318_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_320_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_322_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_325_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_327_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_329_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_331_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_334_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_337_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_339_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_341_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_344_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_346_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_348_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_351_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_353_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_356_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_358_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_361_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_363_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_366_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_368_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_371_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_373_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_375_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_378_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_380_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_383_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_385_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_388_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_390_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_393_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_395_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_398_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_400_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_403_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_405_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_408_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_410_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_413_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_415_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_418_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_420_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_422_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_425_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_427_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_430_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_432_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_435_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_437_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_440_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_442_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_445_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_447_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_450_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_452_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_455_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_457_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_460_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_462_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_465_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_467_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_470_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_472_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_475_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_477_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_480_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_482_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_485_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_487_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_490_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_492_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_495_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_497_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_500_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_502_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_505_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_507_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_510_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_512_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_515_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_517_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_520_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_522_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_525_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_527_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_530_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_532_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_535_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_537_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_540_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_542_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_545_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_547_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_550_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_553_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_555_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_558_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_560_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_563_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_565_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_568_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_570_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_573_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_575_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_578_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_580_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_583_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_586_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_588_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_591_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_593_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_596_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_598_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_601_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_603_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_605_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_608_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_610_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_613_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_615_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_618_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_620_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_623_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_625_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_627_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_630_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_632_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_635_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_637_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_640_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_641_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_642_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_643_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_645_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_646_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_647_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_648_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_650_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_651_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_652_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_653_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_655_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_656_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_657_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_658_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_660_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_661_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_662_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_663_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_665_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_666_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_667_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_668_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_670_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_671_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_672_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_673_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_675_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_676_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_677_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_678_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_679_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_681_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_682_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_683_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_684_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_686_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_687_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_688_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_689_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_691_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_692_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_693_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_694_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_696_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_697_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_698_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_699_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_701_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_702_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_703_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_704_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_706_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_707_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_708_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_709_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_711_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_712_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_713_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_714_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_717_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_719_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_722_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_724_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_727_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_729_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_732_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_734_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_737_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_739_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_741_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_742_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_743_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_744_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_746_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_747_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_748_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_749_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_751_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_752_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_753_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_754_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_756_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_757_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_758_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_759_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_761_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_762_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_763_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_764_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_766_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_767_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_768_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_769_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_771_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_772_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_773_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_774_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_777_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_779_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_782_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_784_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_787_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_789_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_792_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_794_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_797_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_799_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_802_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_804_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_807_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_809_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_812_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_814_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_817_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_819_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_822_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_824_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_827_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_829_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_832_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_835_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_837_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_840_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_842_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_845_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_847_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_850_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_852_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_855_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_857_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_860_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_862_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_865_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_867_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_870_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_872_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_875_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_877_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_880_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_882_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_885_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_887_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_890_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_892_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_895_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_940_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_1038_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_1249_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_1378_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_1618_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_2131_iqu.nc
Loading Rayleigh LUT /opt/ocssw/share/oci/rayleigh/rayleigh_oci_2258_iqu.nc
Using 869.6 nm channel for cloud flagging over water.
Using 412.6 nm channel for cloud flagging over land.
Reading uncertainty from: /opt/ocssw/share/oci/uncertainty.nc
Processing scan # 0 (1 of 1710) after 9 seconds
Aerosol selection bands 751 and 870
NIR correction enabled --> for spectral matching.
Loading aerosol models from /opt/ocssw/share/oci/aerosol/aerosol_oci
Number of Wavelengths 286
Number of Solar Zenith Angles 45
Number of View Zenith Angles 41
Number of Relative Azimuth Angles 20
Number of Principal Components 30
Number of tau 870 Angles 9
Limiting aerosol models based on RH.
Using Spectral Matching of aerosols reflectance for
wavelength from 750.5 nm to 869.6 nm for model selection
Allocated 14633960 bytes in error record.
Allocated 14633960 bytes in error record.
80 aerosol models: 8 humidities x 10 size fractions
model 0, rh=30.000002, sd=5, alpha=2.195868, name=r30f95
model 1, rh=30.000002, sd=20, alpha=2.080902, name=r30f80
model 2, rh=30.000002, sd=50, alpha=1.741297, name=r30f50
model 3, rh=30.000002, sd=70, alpha=1.353765, name=r30f30
model 4, rh=30.000002, sd=80, alpha=1.055717, name=r30f20
model 5, rh=30.000002, sd=90, alpha=0.613691, name=r30f10
model 6, rh=30.000002, sd=95, alpha=0.291905, name=r30f05
model 7, rh=30.000002, sd=98, alpha=0.041746, name=r30f02
model 8, rh=30.000002, sd=99, alpha=-0.055294, name=r30f01
model 9, rh=30.000002, sd=100, alpha=-0.160986, name=r30f00
model 10, rh=50.000000, sd=5, alpha=2.185652, name=r50f95
model 11, rh=50.000000, sd=20, alpha=2.072113, name=r50f80
model 12, rh=50.000000, sd=50, alpha=1.736037, name=r50f50
model 13, rh=50.000000, sd=70, alpha=1.351452, name=r50f30
model 14, rh=50.000000, sd=80, alpha=1.055006, name=r50f20
model 15, rh=50.000000, sd=90, alpha=0.614437, name=r50f10
model 16, rh=50.000000, sd=95, alpha=0.293084, name=r50f05
model 17, rh=50.000000, sd=98, alpha=0.042913, name=r50f02
model 18, rh=50.000000, sd=99, alpha=-0.054214, name=r50f01
model 19, rh=50.000000, sd=100, alpha=-0.160053, name=r50f00
model 20, rh=70.000000, sd=5, alpha=2.162975, name=r70f95
model 21, rh=70.000000, sd=20, alpha=2.066226, name=r70f80
model 22, rh=70.000000, sd=50, alpha=1.770589, name=r70f50
model 23, rh=70.000000, sd=70, alpha=1.416016, name=r70f30
model 24, rh=70.000000, sd=80, alpha=1.131202, name=r70f20
model 25, rh=70.000000, sd=90, alpha=0.689151, name=r70f10
model 26, rh=70.000000, sd=95, alpha=0.351549, name=r70f05
model 27, rh=70.000000, sd=98, alpha=0.078747, name=r70f02
model 28, rh=70.000000, sd=99, alpha=-0.029792, name=r70f01
model 29, rh=70.000000, sd=100, alpha=-0.149895, name=r70f00
model 30, rh=75.000000, sd=5, alpha=2.118432, name=r75f95
model 31, rh=75.000000, sd=20, alpha=2.038649, name=r75f80
model 32, rh=75.000000, sd=50, alpha=1.786870, name=r75f50
model 33, rh=75.000000, sd=70, alpha=1.469576, name=r75f30
model 34, rh=75.000000, sd=80, alpha=1.202941, name=r75f20
model 35, rh=75.000000, sd=90, alpha=0.768258, name=r75f10
model 36, rh=75.000000, sd=95, alpha=0.418238, name=r75f05
model 37, rh=75.000000, sd=98, alpha=0.122661, name=r75f02
model 38, rh=75.000000, sd=99, alpha=0.001526, name=r75f01
model 39, rh=75.000000, sd=100, alpha=-0.135091, name=r75f00
model 40, rh=80.000000, sd=5, alpha=2.023311, name=r80f95
model 41, rh=80.000000, sd=20, alpha=1.955203, name=r80f80
model 42, rh=80.000000, sd=50, alpha=1.735948, name=r80f50
model 43, rh=80.000000, sd=70, alpha=1.450873, name=r80f30
model 44, rh=80.000000, sd=80, alpha=1.204292, name=r80f20
model 45, rh=80.000000, sd=90, alpha=0.789448, name=r80f10
model 46, rh=80.000000, sd=95, alpha=0.444207, name=r80f05
model 47, rh=80.000000, sd=98, alpha=0.144784, name=r80f02
model 48, rh=80.000000, sd=99, alpha=0.019897, name=r80f01
model 49, rh=80.000000, sd=100, alpha=-0.122552, name=r80f00
model 50, rh=85.000000, sd=5, alpha=1.941783, name=r85f95
model 51, rh=85.000000, sd=20, alpha=1.880749, name=r85f80
model 52, rh=85.000000, sd=50, alpha=1.681969, name=r85f50
model 53, rh=85.000000, sd=70, alpha=1.418650, name=r85f30
model 54, rh=85.000000, sd=80, alpha=1.186841, name=r85f20
model 55, rh=85.000000, sd=90, alpha=0.789202, name=r85f10
model 56, rh=85.000000, sd=95, alpha=0.451528, name=r85f05
model 57, rh=85.000000, sd=98, alpha=0.153919, name=r85f02
model 58, rh=85.000000, sd=99, alpha=0.028486, name=r85f01
model 59, rh=85.000000, sd=100, alpha=-0.115537, name=r85f00
model 60, rh=90.000000, sd=5, alpha=1.860837, name=r90f95
model 61, rh=90.000000, sd=20, alpha=1.808119, name=r90f80
model 62, rh=90.000000, sd=50, alpha=1.633757, name=r90f50
model 63, rh=90.000000, sd=70, alpha=1.396786, name=r90f30
model 64, rh=90.000000, sd=80, alpha=1.182832, name=r90f20
model 65, rh=90.000000, sd=90, alpha=0.804794, name=r90f10
model 66, rh=90.000000, sd=95, alpha=0.473155, name=r90f05
model 67, rh=90.000000, sd=98, alpha=0.172704, name=r90f02
model 68, rh=90.000000, sd=99, alpha=0.043690, name=r90f01
model 69, rh=90.000000, sd=100, alpha=-0.106277, name=r90f00
model 70, rh=95.000000, sd=5, alpha=1.741890, name=r95f95
model 71, rh=95.000000, sd=20, alpha=1.702224, name=r95f80
model 72, rh=95.000000, sd=50, alpha=1.567437, name=r95f50
model 73, rh=95.000000, sd=70, alpha=1.375453, name=r95f30
model 74, rh=95.000000, sd=80, alpha=1.193434, name=r95f20
model 75, rh=95.000000, sd=90, alpha=0.851522, name=r95f10
model 76, rh=95.000000, sd=95, alpha=0.529470, name=r95f05
model 77, rh=95.000000, sd=98, alpha=0.218423, name=r95f02
model 78, rh=95.000000, sd=99, alpha=0.078690, name=r95f01
model 79, rh=95.000000, sd=100, alpha=-0.088852, name=r95f00
chl_hu: using 442.33 547.44 669.52
rh_ndims=5 rh_dimids=4 3 2 1 0
morel f/q file dimensions n_a=13 n_n=17 n_c=6 n_s=6 n_w=7
Reading foq file /opt/ocssw/share/common/morel_fq.nc ndims=5 nvars=6 sds_id=5 var=foq
Closing foq file /opt/ocssw/share/common/morel_fq.nc
Morel f/Q table from file /opt/ocssw/share/common/morel_fq.nc
Applying ocean BRDF including:
Reflection/refraction for upwelling radiance.
Reflection/refraction for downwelling radiance.
Morel f/Q
Compute Raman scattering correction #2.
Loading Raman coefficients from: /opt/ocssw/share/oci/raman.nc.
Warning: Cannot apply GMAO total extinction absorbing aerosol test without
anc_aerosol inputs defined
Setting absaer_opt=0
Reading pca table for picophyto from: /opt/ocssw/share/common/pca_picophyto.h5
Processing scan # 50 (51 of 1710) after 112 seconds
Processing scan # 100 (101 of 1710) after 216 seconds
Processing scan # 150 (151 of 1710) after 318 seconds
Processing scan # 200 (201 of 1710) after 426 seconds
Processing scan # 250 (251 of 1710) after 540 seconds
Processing scan # 300 (301 of 1710) after 659 seconds
Processing scan # 350 (351 of 1710) after 790 seconds
Processing scan # 400 (401 of 1710) after 938 seconds
Processing scan # 450 (451 of 1710) after 1097 seconds
Processing scan # 500 (501 of 1710) after 1270 seconds
Processing scan # 550 (551 of 1710) after 1385 seconds
Processing scan # 600 (601 of 1710) after 1469 seconds
Processing scan # 650 (651 of 1710) after 1548 seconds
Processing scan # 700 (701 of 1710) after 1627 seconds
Processing scan # 750 (751 of 1710) after 1707 seconds
Processing scan # 800 (801 of 1710) after 1791 seconds
Processing scan # 850 (851 of 1710) after 1868 seconds
Processing scan # 900 (901 of 1710) after 1936 seconds
Processing scan # 950 (951 of 1710) after 2005 seconds
Processing scan # 1000 (1001 of 1710) after 2071 seconds
Processing scan # 1050 (1051 of 1710) after 2137 seconds
Processing scan # 1100 (1101 of 1710) after 2206 seconds
Processing scan # 1150 (1151 of 1710) after 2274 seconds
Processing scan # 1200 (1201 of 1710) after 2337 seconds
Processing scan # 1250 (1251 of 1710) after 2397 seconds
Processing scan # 1300 (1301 of 1710) after 2455 seconds
Processing scan # 1350 (1351 of 1710) after 2510 seconds
Processing scan # 1400 (1401 of 1710) after 2557 seconds
Processing scan # 1450 (1451 of 1710) after 2594 seconds
Processing scan # 1500 (1501 of 1710) after 2630 seconds
Processing scan # 1550 (1551 of 1710) after 2658 seconds
Processing scan # 1600 (1601 of 1710) after 2685 seconds
Processing scan # 1650 (1651 of 1710) after 2713 seconds
Processing scan # 1700 (1701 of 1710) after 2744 seconds
Percentage of pixels flagged:
Flag # 1: ATMFAIL 453 0.0208
Flag # 2: LAND 973253 44.7448
Flag # 3: PRODWARN 0 0.0000
Flag # 4: HIGLINT 0 0.0000
Flag # 5: HILT 43466 1.9983
Flag # 6: HISATZEN 312761 14.3790
Flag # 7: COASTZ 5152 0.2369
Flag # 8: SPARE 0 0.0000
Flag # 9: STRAYLIGHT 244154 11.2249
Flag #10: CLDICE 766199 35.2256
Flag #11: COCCOLITH 2166 0.0996
Flag #12: TURBIDW 1944 0.0894
Flag #13: HISOLZEN 0 0.0000
Flag #14: SPARE 0 0.0000
Flag #15: LOWLW 2511 0.1154
Flag #16: CHLFAIL 646 0.0297
Flag #17: NAVWARN 0 0.0000
Flag #18: ABSAER 0 0.0000
Flag #19: SPARE 0 0.0000
Flag #20: MAXAERITER 1023 0.0470
Flag #21: MODGLINT 0 0.0000
Flag #22: CHLWARN 172 0.0079
Flag #23: ATMWARN 1114 0.0512
Flag #24: SPARE 2738 0.1259
Flag #25: SEAICE 0 0.0000
Flag #26: NAVFAIL 0 0.0000
Flag #27: FILTER 0 0.0000
Flag #28: SPARE 0 0.0000
Flag #29: BOWTIEDEL 0 0.0000
Flag #30: HIPOL 0 0.0000
Flag #31: PRODFAIL 1514553 69.6308
Flag #32: SPARE 0 0.0000
End MSl12 processing at 2024217035110000
Processing Rate = 0.621818 scans/sec
Closing oci l1b file
Processing Completed
dataset = xr.open_dataset(par["ofile"], group="geophysical_data")
artist = dataset["picoeuk_moana"].plot(cmap="viridis", robust=True)
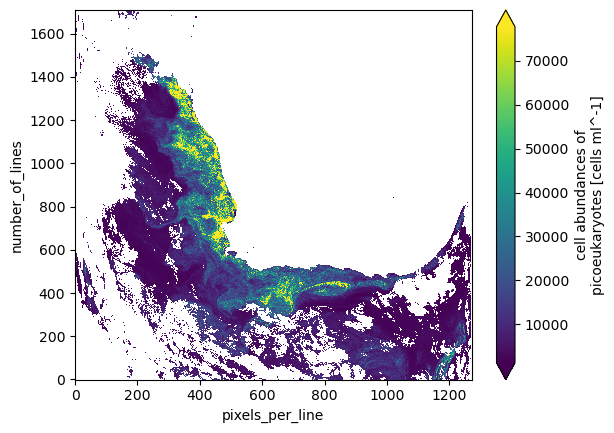
5. Composite L2 Data with l2bin#
It can be useful to merge adjacent scenes to create a single, larger image. Let’s do it with a chlorophyll product.
Searching on a location defined as a line, rather than a point, is a good way to get granules that are
adjacent to eachother. Pass a list of latitude and longitude tuples to the line argument of search_data.
tspan = ("2024-04-27", "2024-04-27")
location = [(-56.5, 49.8), (-55.0, 49.8)]
results = earthaccess.search_data(
short_name="PACE_OCI_L2_BGC_NRT",
temporal=tspan,
line=location,
)
for item in results:
display(item)
paths = earthaccess.download(results, "data")
While we have the downloaded location stored in the list paths, write the list to a text file for future use.
paths = [str(i) for i in paths]
with open("l2bin_ifile.txt", "w") as file:
file.write("\n".join(paths))
The OCSSW program that performs merging, also known as “compositing” of remote sensing images, is called l2bin. Take a look at the program’s options.
%%bash
source $OCSSWROOT/OCSSW_bash.env
l2bin help
l2bin 7.0.7 (Jul 17 2024 13:15:06)
7.0.7
Usage: l2bin argument-list
The argument-list is a set of keyword=value pairs. The arguments can
be specified on the commandline, or put into a parameter file, or the
two methods can be used together, with commandline over-riding.
return value: 0=OK, 1=error, 110=north,south,east,west does not intersect
file data.
The list of valid keywords follows:
help (boolean) (alias=h) (default=false) = print usage information
version (boolean) (default=false) = print the version
information
dump_options (boolean) (default=false) = print
information about each option
dump_options_paramfile (ofile) = print
information about each option to paramfile
dump_options_xmlfile (ofile) = print
information about each option to XML file
par (ifile) (alias=parfile) = input parameter file
ifile (ifile) (alias=infile) = input L2 file name
ofile (ofile) (default=output) = output file name
fileuse (ofile) = write filenames of the input files used into this file
suite (string) = suite for default parameters
qual_prod (string) = quality product field name
deflate (int) (default=5) = deflation level. 0=off or 1=low through 9=high
verbose (boolean) (default=off) = Allow more verbose screen messages
night (boolean) (default=off) = set to 1 for SST night processing
qual_max (int) (default=2) = maximum acceptable quality
rowgroup (int) (default=-1) = # of bin rows to process at once.
sday (int) (default=1970001) = start datadate (YYYYDDD) [ignored for "regional" prodtype]
eday (int) (default=2038018) = end datadate (YYYYDDD) [ignored for "regional" prodtype]
latnorth (float) (default=90) = northern most latitude
latsouth (float) (default=-90) = southern most latitude
loneast (float) (default=0) = eastern most longitude
lonwest (float) (default=0) = western most longitude
minobs (int) (default=0) = required minimum number of observations
delta_crossing_time (float) (default=0.0) = equator crossing time delta in
minutes
Caveat...if zero, the sensor default equator crossing time will be used
This is not necessarily noon
resolution (string) (alias=resolve) (default=H) = bin resolution
H: 0.5km
Q: 250m
HQ: 100m
HH: 50m
1: 1.1km
2: 2.3km
4: 4.6km
9: 9.2km
18: 18.5km
36: 36km
1D: 1 degree
HD: 0.5 degree
QD: 0.25 degree
prodtype (string) (default=day) = product type (Set to "regional" to bin all scans.)
pversion (string) (default=unspecified) = production version
composite_scheme (string) = composite scheme (min/max)
composite_prod (string) = composite product fieldname
flaguse (string) (default=ATMFAIL,LAND,HILT,HISATZEN,STRAYLIGHT,CLDICE,COCCOLITH,LOWLW,CHLFAIL,CHLWARN,NAVWARN,ABSAER,MAXAERITER,ATMWARN,HISOLZEN,NAVFAIL,FILTER) = flags masked [see /SENSOR/l2bin_defaults.par]
l3bprod (string) (default=ALL) = l3bprod = bin products [default=all products]
Set to "ALL" or "all" for all L2 products in 1st input file.
Use ':' or ',' or ' ' as delimiters.
Use ';' or '=' to delineate minimum values.
area_weighting (int) (default=0) = Enable area weighting
0: off
1: pixel box
2: pixel bounding box
3: pixel polygon
output_wavelengths (string) (default=ALL) = comma separated list of
wavelengths for multi-wavelength products
doi (string) = Digital Object Identifier (DOI) string
oprodname (string) = comma separated list of output L3 product names.
This option allows the user to specify the output product names which differ from the original l2 product names.
Usage: original_l2_name:output_l3_name, i.e. oprodname=cloud_flag:cloud_fraction
Write a parameter file with the previously saved list of Level-2 BGC files standing in for the usual “ifile” value. We can leave the datetime out of the “ofile” name rather than extracing a time period from the granules chosen for binning.
ofile = "data/PACE_OCI.L3B.nc"
par = {
"ifile": "l2bin_ifile.txt",
"ofile": ofile,
"prodtype": "regional",
"resolution": 9,
"flaguse": "NONE",
"rowgroup": 2000,
}
write_par("l2bin.par", par)
%%bash
source $OCSSWROOT/OCSSW_bash.env
l2bin par=l2bin.par
l2bin 7.0.7 (Jul 17 2024 13:15:06)
Loading default parameters from /opt/ocssw/share/common/l2bin_defaults.par
Loading default parameters from /opt/ocssw/share/oci/l2bin_defaults.par
Loading command line parameters
2 input files
Resolution: 9
Max Qual Allowed: 255
total number of bins: 5940422
Input row_group: 2000 Actual row_group: 1080
krow: 0 out of 2160 (-90.00 to 0.00) Sun Aug 4 03:51:19 2024
krow: 1080 out of 2160 ( 0.00 to 90.00) Sun Aug 4 03:51:19 2024
# bins in row group : 2970211
# filled bins : 29249
total_filled_bins: 29249
dataset = xr.open_dataset(par["ofile"], group="level-3_binned_data")
dataset
<xarray.Dataset> Size: 2MB
Dimensions: (binListDim: 29249, binDataDim: 29249, binIndexDim: 2160)
Dimensions without coordinates: binListDim, binDataDim, binIndexDim
Data variables:
BinList (binListDim) {'names': ['bin_num', 'nobs', 'nscenes', 'weights', 'time_rec'], 'formats': ['<u4', '<i2', '<i2', '<f4', '<f4'], 'offsets': [0, 4, 6, 8, 12], 'itemsize': 16, 'aligned': True} 468kB ...
chlor_a (binDataDim) {'names': ['sum', 'sum_squared'], 'formats': ['<f4', '<f4'], 'offsets': [0, 4], 'itemsize': 8, 'aligned': True} 234kB ...
carbon_phyto (binDataDim) {'names': ['sum', 'sum_squared'], 'formats': ['<f4', '<f4'], 'offsets': [0, 4], 'itemsize': 8, 'aligned': True} 234kB ...
poc (binDataDim) {'names': ['sum', 'sum_squared'], 'formats': ['<f4', '<f4'], 'offsets': [0, 4], 'itemsize': 8, 'aligned': True} 234kB ...
chlor_a_unc (binDataDim) {'names': ['sum', 'sum_squared'], 'formats': ['<f4', '<f4'], 'offsets': [0, 4], 'itemsize': 8, 'aligned': True} 234kB ...
carbon_phyto_unc (binDataDim) {'names': ['sum', 'sum_squared'], 'formats': ['<f4', '<f4'], 'offsets': [0, 4], 'itemsize': 8, 'aligned': True} 234kB ...
BinIndex (binIndexDim) {'names': ['start_num', 'begin', 'extent', 'max'], 'formats': ['<u4', '<u4', '<u4', '<u4'], 'offsets': [0, 4, 8, 12], 'itemsize': 16, 'aligned': True} 35kB ...5. Make a Map from Binned Data with l3mapgen#
The l3mapgen function of OCSSW allows you to create maps with a wide array of options you can see below:
%%bash
source $OCSSWROOT/OCSSW_bash.env
l3mapgen help
l3mapgen 2.4.0-V2024.1 (Jul 17 2024 13:14:38)
Usage: l3mapgen argument-list
This program takes a product (or products if netCDF output) from an L3 bin
or SMI file, reprojects the data using proj.4 and writes a mapped file in
the requested output format.
Return values
0 = All Good
1 = Error
110 = No valid data to map
The argument list is a set of keyword=value pairs. Arguments can
be specified on the command line, or put into a parameter file, or the
two methods can be used together, with command line overriding.
The list of valid keywords follows:
help (boolean) (alias=h) (default=false) = print usage information
version (boolean) (default=false) = print the version
information
dump_options (boolean) (default=false) = print
information about each option
dump_options_paramfile (ofile) = print
information about each option to paramfile
dump_options_xmlfile (ofile) = print
information about each option to XML file
par (ifile) = input parameter file
suite (string) = suite for default parameters
ifile (ifile) = input L3 bin filename
ofile (ofile) (default=output) = output filename
oformat (string) (default=netcdf4) = output file format
netcdf4: netCDF4 file, can contain more than one product
hdf4: HDF4 file (old SMI format)
png: PNG image file
ppm: PPM image file
tiff: TIFF file with georeference tags
ofile_product_tag (string) (default=PRODUCT) = sub-string in ofile name that will be substituted by the product name
ofile2 (ofile) = second output filename
oformat2 (string) (default=png) = second output file format
same options as oformat
deflate (int) (default=4) = netCDF4 deflation level
product (string) = comma separated list of products.
Each product can have an optional colon and modifier appended.
For example, "product=chlor_a,chlor_a:stdev,Kd_490:nobs"
Available modifiers:
avg average value (default)
stdev standard deviation
var variance
nobs number of observations in the bin
nscenes number of contributing scenes
obs_time average observation time (TAI93)
bin_num bin ID number
wavelength_3d (string) = comma separated list of wavelengths for 3D products
resolution (string) = size of output pixel (default from input file)
in meters or SMI dimensions
90km: 432 x 216 image for full globe
36km: 1080 x 540
18km: 2160 x 1080
9km: 4320 x 2160
4km: 8640 x 4320
2km: 17280 x 8640
1km: 34560 x 17280
hkm: 69120 x 34560
qkm: 138240 x 69120
smi: 4096 x 2048
smi4: 8192 x 4096
land: 8640 x 4320
#.#: width of a pixel in meters
#.#km: width of a pixel in kilometers
#.#deg: width of a pixel in degrees
width (int) = width of output image in pixels; supercedes resolution parameter.
projection (string) (default=platecarree) = proj.4 projection string or one
of the following predefined projections:
smi: Standard Mapped image, cylindrical projection,
uses central_meridian. NSEW defaults to whole globe.
projection="+proj=eqc +lon_0=<central_meridian>"
platecarree: Plate Carree image, cylindrical projection,
uses central_meridian.
projection="+proj=eqc +lon_0=<central_meridian>"
mollweide: Mollweide projection
projection="+proj=moll +lon_0=<central_meridian>"
lambert: Lambert conformal conic (2SP) projection
projection="+proj=lcc +lon_0=<central_meridian>
+lat_0=<scene center latitude>
+lat_1=<scene south latitude>
+lat_2=<scene north latitude>"
albersconic: Albers Equal Area Conic projection
projection="+proj=aea +lon_0=<central_meridian>
+lat_0=<scene center latitude>
+lat_1=<scene south latitude>
+lat_2=<scene north latitude>"
aeqd: Azimuthal Equidistant projection
projection="+proj=aeqd +lon_0=<central_meridian>
+lat_0=<scene center latitude>"
mercator: Mercator cylindrical map projection
projection="+proj=merc +lon_0=<central_meridian>"
transmerc: Transverse Mercator cylindrical map projection
projection="+proj=tmerc +lon_0=<central_meridian>
+lat_0=<scene center latitude>"
utm: Universal Transverse Mercator cylindrical map projection
projection="+proj=utm +zone=<utm_zone> [+south]"
obliquemerc: Oblique Mercator cylindrical map projection
projection="+proj=omerc +gamma=0 +lat_0=<lat_0>
+lonc=<central_meridian> +alpha=<azimuth>
+k_0=1 +x_0=0 +y_0=0"
ease2: EASE-Grid 2.0 projection
projection="EPSG:6933"
ortho: Orthographic projection
projection="+proj=ortho +lat_0=<lat_0> +lon_0=<central_meridian>
+ellps=GRS80 +units=m"
stere: Stereographic projection
projection="+proj=stere +lat_0=<lat_0> +lat_ts=<lat_ts>
+lon_0=<central_meridian>
+ellps=WGS84 +datum=WGS84 +units=m"
conus: USA Contiguous Albers Equal Area Conic USGS version
projection="+proj=aea +lat_1=29.5 +lat_2=45.5
+lat_0=23.0 +lon_0=-96 +x_0=0 +y_0=0
+ellps=GRS80 +datum=NAD83 +units=m"
alaska: Alaskan Albers Equal Area Conic USGS version
projection="EPSG:3338"
gibs: latitudinally dependent projection
Plate Carree between 60S and 60N
else use Polar Sterographic
North Polar: projection="EPSG:3413"
South Polar: projection="EPSG:3031"
raw: Raw dump of bin file contents.
write_projtext (boolean) (default=no) = write projection information to a text file.
central_meridian (float) (default=-999) = central meridian for projection in deg east.
Used only for raw dump and predefined projections as above.
lat_ts (float) = latitude of true scale for projection in deg north.
Used only for predefined projections above as required.
lat_0 (float) = latitude of origin for projection in deg north.
Used only for predefined projections above as required.
lat_1 (float) = latitude of first standard parallel (south).
Used only for predefined projections above as required.
lat_2 (float) = latitude of second standard parallel (north).
Used only for predefined projections above as required.
azimuth (float) = projection rotation angle in deg north.
Used only for predefined projections above as required.
utm_zone (string) = UTM zone number.
Used only for the UTM projection;
Append 'S' for southern hemisphere zones (e.g. 59S).
north (float) (default=-999) = Northernmost Latitude (default: file north)
south (float) (default=-999) = Southernmost Latitude (default: file south)
east (float) (default=-999) = Easternmost Longitude (default: file east)
west (float) (default=-999) = Westernmost Longitude (default: file west)
trimNSEW (boolean) (default=no) = should we trim output
to match input NSEW range
interp (string) (default=nearest) = interpolation method:
nearest: use the value of the nearest bin for the pixel
bin: bin all of the pixels that intersect the area of the
output pixel
area: bin weighted by area of all the pixels that intersect
the area of the output pixel
apply_pal (boolean) (default=yes) = apply color palette:
yes: color image
no: grayscale image
palfile (ifile) = palette filename (default from product.xml)
use_transparency (boolean) (default=no) = make missing data transparent (only valid for color PNG and TIFF)
datamin (float) = minimum value for scaling (default from product.xml)
datamax (float) = maximum value for scaling (default from product.xml)
scale_type (string) = data scaling type (default from product.xml)
linear: linear scaling
log: logarithmic scaling
arctan: arc tangent scaling
quiet (boolean) (default=false) = stop the status printing
pversion (string) (default=Unspecified) = processing version string
use_quality (boolean) (default=yes) = should we do quality factor processing
quality_product (string) = product to use for quality factor processing
use_rgb (boolean) (default=no) = should we use product_rgb to make a
pseudo-true color image
product_rgb (string) (default=rhos_670,rhos_555,rhos_412) =
Three products to use for RGB. Default is sensor-specific.
fudge (float) (default=1.0) = fudge factor used to modify size of L3 pixels
threshold (float) (default=0) = minimum percentage of filled pixels before
an image is generated
num_cache (int) (default=500) = number of rows to cache in memory.
mask_land (boolean) (default=no) = set land pixels to pixel value 254
rgb_land (string) (default=160,82,45) = RGB value to use for land mask; comma separate string
land (ifile) (default=$OCDATAROOT/common/landmask_GMT15ARC.nc) = land mask file
full_latlon (boolean) (default=yes) = write full latitude and longitude arrays.
doi (string) = Digital Object Identifier (DOI) string
Run l3mapgen to make a 9km map with a Plate Carree projection.
ifile = "data/PACE_OCI.L3B.nc"
ofile = ifile.replace(".L3B.", ".L3M.")
par = {
"ifile": ifile,
"ofile": ofile,
"projection": "platecarree",
"resolution": "9km",
"interp": "bin",
"use_quality": 0,
"apply_pal": 0,
}
write_par("l3mapgen.par", par)
%%bash
source $OCSSWROOT/OCSSW_bash.env
l3mapgen par=l3mapgen.par
Loading default parameters from /opt/ocssw/share/oci/l3mapgen_defaults.par
l3mapgen 2.4.0-V2024.1 (Jul 17 2024 13:14:38)
ifile : data/PACE_OCI.L3B.nc
ofile : data/PACE_OCI.L3M.nc
oformat : netcdf4
projection : platecarree
resolution : 9276.624m
product : chlor_a,carbon_phyto,poc,chlor_a_unc,carbon_phyto_unc
north : 57.708
south : 35.458
east : -15.967
west : -72.537
image size : 269 x 681
99% complete
actual data min : 0.001000
actual data max : 1996.799805
num filled pixels : 48565
percent filled pixels : 26.51%
Open the output with XArray, note that there is no group anymore and that lat and lon are coordinates as well as indexes.
dataset = xr.open_dataset(par["ofile"])
dataset
<xarray.Dataset> Size: 4MB
Dimensions: (lat: 269, lon: 681, rgb: 3, eightbitcolor: 256)
Coordinates:
* lat (lat) float32 1kB 57.67 57.58 57.5 ... 35.67 35.58 35.5
* lon (lon) float32 3kB -72.49 -72.41 -72.33 ... -16.09 -16.01
Dimensions without coordinates: rgb, eightbitcolor
Data variables:
chlor_a (lat, lon) float32 733kB ...
carbon_phyto (lat, lon) float32 733kB ...
poc (lat, lon) float32 733kB ...
chlor_a_unc (lat, lon) float32 733kB ...
carbon_phyto_unc (lat, lon) float32 733kB ...
palette (rgb, eightbitcolor) uint8 768B ...
Attributes: (12/61)
product_name: PACE_OCI.L3M.nc
instrument: OCI
title: OCI Level-3 Equidistant Cylindrical Map...
project: Ocean Biology Processing Group (NASA/GS...
platform: PACE
source: satellite observations from OCI-PACE
... ...
processing_level: L3 Mapped
cdm_data_type: grid
proj4_string: +proj=eqc +lat_ts=0 +lat_0=0 +x_0=0 +y_...
data_bins: 48565
data_minimum: 0.0009999998
data_maximum: 1996.7998Now that we have projected chlorophyll data, we can make a map with coastines and gridlines provided by Cartopy. The projection argument to plt.axes indicates the projection we want to have in the visualization. The transform
argument in the Dataset.plot call indicates the projection the data are in. Cartopy does reprojection easilly
between two projected coordinate reference systems.
fig = plt.figure(figsize=(10, 3))
ax = plt.axes(projection=ccrs.AlbersEqualArea(-45))
artist = dataset["chlor_a"].plot(x="lon", y="lat", cmap="viridis", robust=True, ax=ax, transform=ccrs.PlateCarree())
ax.gridlines(draw_labels={"left": "y", "bottom": "x"}, linewidth=0.2)
ax.coastlines(linewidth=0.5)
plt.show()